(5) A Predator-Prey Model
Dustin Kapraun
2026-03-01
Source:vignettes/pred_prey_demo.Rmd
pred_prey_demo.RmdModel Description
Suppose there is a population of rabbits (prey) and a population of
foxes (predators) that inhabit the same area. The sizes of these two
populations can be modeled as a system of ordinary differential
equations (ODEs). In particular, if
and
represent the numbers (in thousands) of rabbits and foxes, respectively,
the rates of change of these numbers can be estimated as
where
(day)
is a parameter that determines the birth rate of rabbits,
(day
per 1000 foxes) is a parameter that determines the death rate of
rabbits,
(day
per 1000 rabbits) is a parameter that determines the birth rate of
foxes, and
(day)
is a parameter that determines the death rate of foxes. In order to
solve an initial value problem for the predator-prey model, one needs to
provide the values of
,
,
,
and
,
as well as initial values for
and
.
MCSim Model Specification
We used the GNU
MCSim model specification language to implement the predator-prey
model. The complete MCSim model specification file for this model,
pred_prey.model, can be found in the extdata
subdirectory of the MCSimMod package installation
directory.
In the model specification file, the text symbols x and
y are used to represent the state variables
and
and the text symbols alpha, beta,
gamma, and delta are used to represent the
parameters
,
,
,
and
,
respectively.
Building the Model
First, we load the MCSimMod package as follows.
Using the following commands, we create a model object (i.e., an
instance of the Model class) using the model specification
file pred_prey.model that is included in the
MCSimMod package.
# Get the full name of the package directory that contains the example MCSim
# model specification file.
mod_path <- file.path(system.file(package = "MCSimMod"), "extdata")
# Create a model object using the example MCSim model specification file
# "pred_prey.model" included in the MCSimMod package.
pp_mod_name <- file.path(mod_path, "pred_prey")
pp_mod <- createModel(pp_mod_name)Once the model object is created, we can “load” the model (so that it’s ready for use in a given R session) as follows.
# Load the model.
pp_mod$loadModel()Predicting the Numbers of Rabbits and Foxes
Suppose we want to predict the numbers of rabbits and foxes over a period of 50 days assuming that , , , , and the initial numbers of rabbits and foxes are 1000 and 750, respectively. These are the default values of the model parameters and initial conditions that are provided in the model specification file, and we can verify this with the following commands.
pp_mod$parms
#> alpha beta gamma delta
#> 0.67 1.33 1.00 1.00
pp_mod$Y0
#> x y
#> 1.00 0.75We can perform a simulation that provides results for the desired output times (i.e., ) using the following commands.
# Define output times for simulation.
times <- seq(from = 0, to = 50, by = 0.1)
# Run simulation.
out <- pp_mod$runModel(times)Examining the Results
The final command shown above,
out <- pp_mod$runModel(times), performs a model
simulation and stores the simulation results in a “matrix” data
structure called out. There is one row for each output
time, and one column for each state variable. The first five rows of
this data structure are shown below. Note that the independent variable,
which is
in the case of the predator-prey model, is always labeled “time” in the
output data structure.
| time | x | y |
|---|---|---|
| 0.0 | 1.0000000 | 0.7500000 |
| 0.1 | 0.9678346 | 0.7487880 |
| 0.2 | 0.9369984 | 0.7452243 |
| 0.3 | 0.9077112 | 0.7394507 |
| 0.4 | 0.8801364 | 0.7316377 |
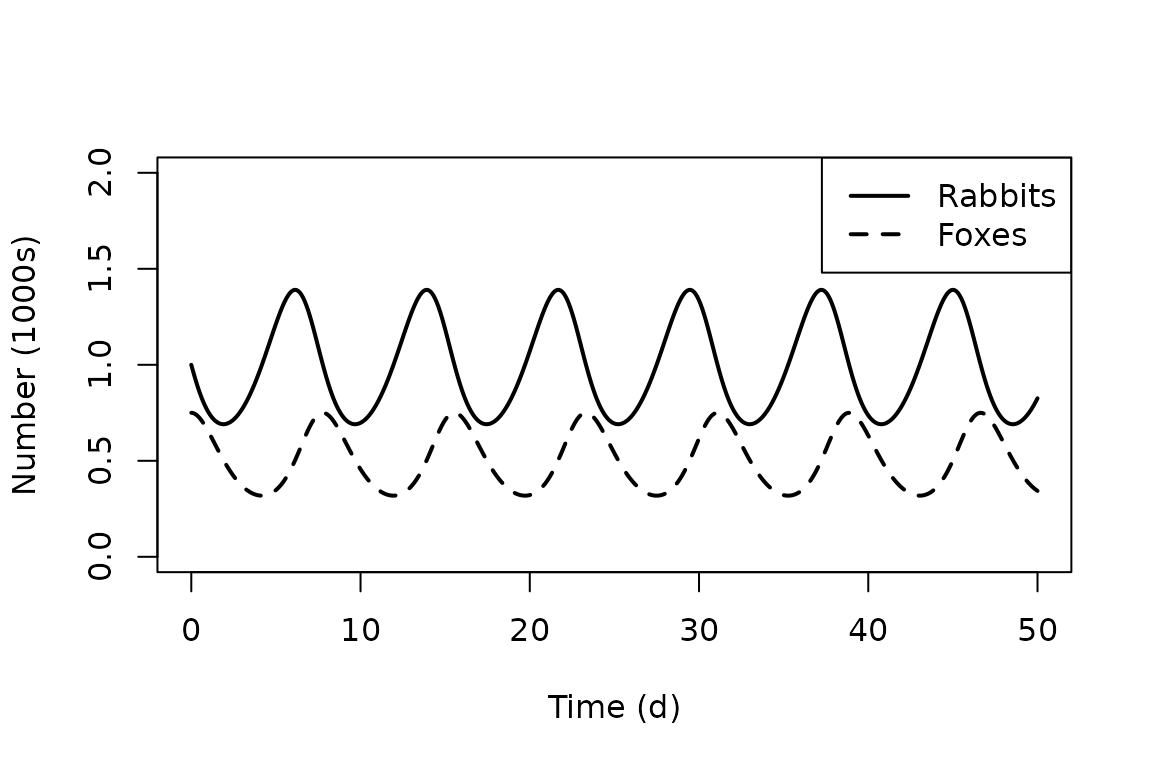
We can create visual representations of the simulation results. For example, we can plot the numbers of rabbits and foxes vs. time using the following commands.
# Plot simulation results (numbers vs. time).
plot(out[, "time"], out[, "x"],
type = "l", lty = 1, lwd = 2,
xlab = "Time (d)", ylab = "Number (1000s)", ylim = c(0, 2)
)
lines(out[, "time"], out[, "y"], lty = 2, lwd = 2)
legend("topright", c("Rabbits", "Foxes"),
lty = c(1, 2),
lwd = 2
)
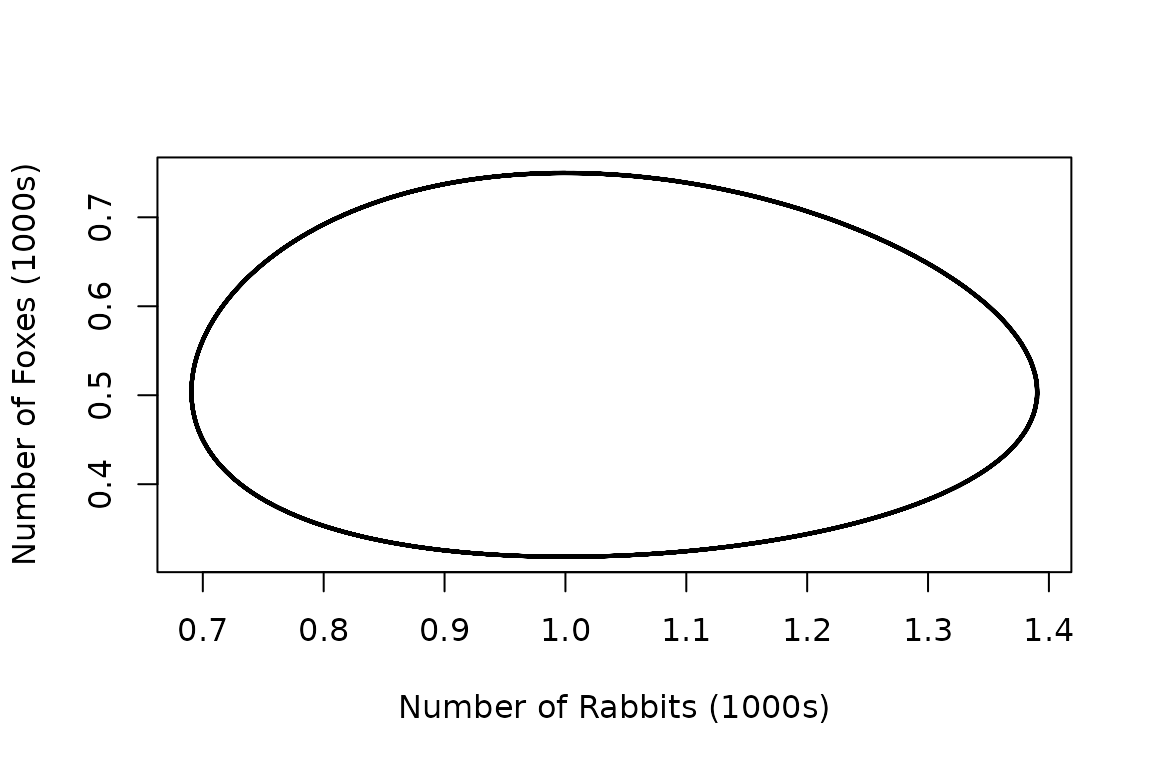
Alternatively, we can plot the results in phase-space as follows.
# Plot simulation results (number of foxes vs. number of rabbits).
plot(out[, "x"], out[, "y"],
type = "l", lty = 1, lwd = 2,
xlab = "Number of Rabbits (1000s)", ylab = "Number of Foxes (1000s)"
)