Plotting watershed data
In this example we access a single variable for the Calapooia River
using sc_get_data function. We then use the
nhdplusTools library to grab flowlines and watershed for
the Calapooia, plot the selected StreamCat metric for the Calapooia
River and show the watershed.
library(StreamCatTools)
start_comid = 23763517
nldi_feature <- list(featureSource = "comid", featureID = start_comid)
flowline_nldi <- nhdplusTools::navigate_nldi(nldi_feature, mode = "UT", data_source = "flowlines", distance=5000)
# get StreamCat metrics
df <- sc_get_data(metric='pctimp2011', aoi='cat', comid=flowline_nldi$UT_flowlines$nhdplus_comid)
# We can also pull out comids the following way:
# comids <- paste(as.integer(flowline_nldi$UT_flowlines$nhdplus_comid), collapse=",",sep="")
# df <- sc_get_data(metric='pctimp2011', aoi='cat', comid=comids)
flowline_nldi <- flowline_nldi$UT_flowlines
flowline_nldi$PCTIMP2011CAT <- df$pctimp2011cat[match(flowline_nldi$nhdplus_comid, df$comid)]
basin <- nhdplusTools::get_nldi_basin(nldi_feature = nldi_feature)
library(mapview)
mapview::mapviewOptions(fgb=FALSE)
mapview::mapview(basin, alpha.regions=.08) + mapview::mapview(flowline_nldi, zcol = "PCTIMP2011CAT", legend = TRUE)Working with NARS data
In this example we demonstrate a data ‘mashup’ by grabbing NRSA data
from the EPA National Aquatic Resource Surveys (NARS) website directly
in R, pull particular StreamCat metrics for sites using
sc_get_data, and compare landscape metrics with other NRSA
metrics
nrsa <- readr::read_csv("https://www.epa.gov/sites/production/files/2015-09/siteinfo_0.csv")
dplyr::glimpse(nrsa)
# Promote data frame to sf spatial points data frame
nrsa_sf <- sf::st_as_sf(nrsa, coords = c("LON_DD83", "LAT_DD83"), crs = 4269)
# Get COMIDs using nhdplusTools package
# nrsa$COMID<- NA
# for (i in 1:nrow(nrsa_sf)){
# print (i)
# nrsa_sf[i,'COMID'] <- discover_nhdplus_id(nrsa_sf[i,c('geometry')])
# }
load(system.file("extdata", "sample_nrsa_data.rda", package="StreamCatTools"))
# get particular StreamCat data for all these NRSA sites
# nrsa_sf$COMID <- as.character(nrsa_sf$COMID)
comids <- nrsa_sf$COMID
comids <- comids[!is.na(comids)]
comids <- comids[c(1:700)]
comids <- paste(comids,collapse=',')
df <- sc_get_data(metric='pctcrop2006', aoi='ws', comid=comids)
# glimpse(df)
df$COMID <- as.integer(df$comid)
nrsa_sf <- dplyr::left_join(nrsa_sf, df, by='COMID')
# download mmi from NARS web page
library(dplyr)
library(ggplot2)
mmi <- readr::read_csv("https://www.epa.gov/sites/production/files/2015-09/bentcond.csv")
# dplyr::glimpse(mmi)
# join mmi to NARS info data frame with StreamCat PctCrop metric
nrsa_sf <- dplyr::left_join(nrsa_sf, mmi[,c('SITE_ID','BENT_MMI_COND')], by='SITE_ID')
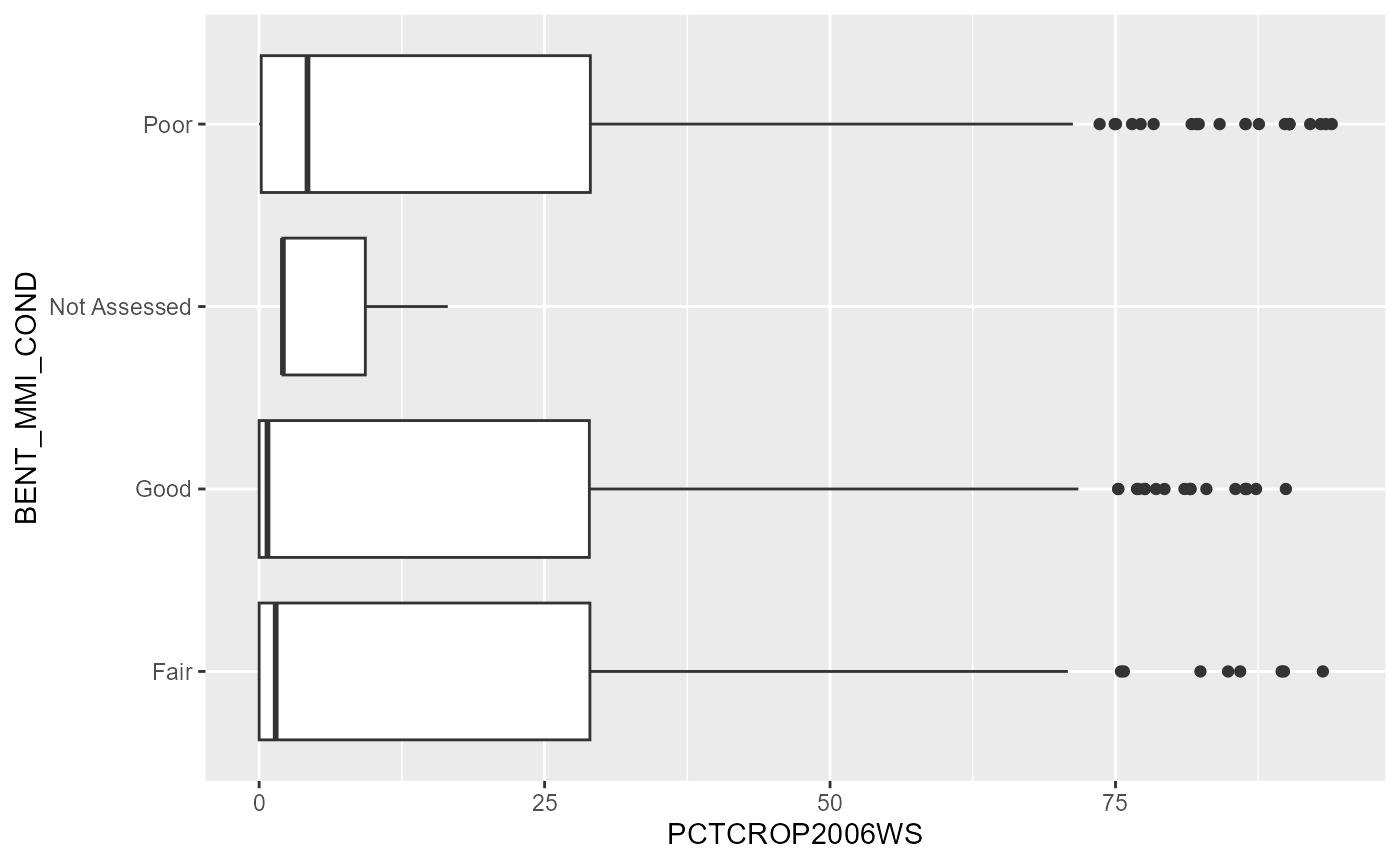
bxplt <- nrsa_sf %>%
tidyr::drop_na(BENT_MMI_COND) %>%
ggplot2::ggplot(aes(x=pctcrop2006ws, y=BENT_MMI_COND))+
ggplot2::geom_boxplot()+
ggplot2::ggtitle('NRSA Benthic MMI versus % Crop in Watershed from 2006 NLCD')
suppressWarnings(print(bxplt))