rExpertQuery Cybertown Training
ATTAINS Team
2025-06-02
CybertownTraining.RmdAbout Expert Query and rExpertQuery
Expert Query empowers users to access surface water quality data from the Assessment and TMDL Tracking and Implementation System (ATTAINS) encompassing Assessment decisions (under Clean Water Act Sections 303(d), 305(b), and 106) and Action data like Total Maximum Daily Loads (TMDLs), Advance Restoration Plans (ARPs), and Protection Approaches.
The tool supports querying and downloading data within and across organizations (states or tribes), such as querying all the data within an EPA region or all nutrient TMDLs nationally. It also allows users to download national ATTAINS data extracts.
rExpertQuery takes this a step further as its functions facilitate querying the Expert Query web services and importing the resulting data sets directly in to R for more detailed review and analysis.
It is crucial to be aware of certain complexities inherent in the data and querying process. Users are cautioned against direct comparisons between states due to differences in water quality standards and assessment methodologies. The summary and count examples provided here are intended to demonstrate potential uses of rExpertQuery and complementary R packages, but do not imply EPA’s endorsement of any specific summary or count method.
While Expert Query facilitates data querying, it lacks built-in summary capabilities. Users are encouraged to download and summarize data, but use caution when relying on row counts, as duplicate entries may occur. This complexity arises from multiple tables that have many-to-many relationships. Several examples of summarizing Expert Query data are included in this vignette. These examples use additional packages for data wrangling, mapping, visualizing and other tasks. Future rExpertQuery vignettes and functions may provide additional examples of how to summarize Expert Query data.
Installation
You must first have R and R Studio installed to use the rExpert Query (see instructions below if needed). rExpert Query is in active development, therefore we highly recommend that you update it and all of its dependency libraries each time you use the package.
You can install and/or update the rExpertQuery Package and all dependencies by running:
# install and load rExpertQuery
if (!"remotes" %in% installed.packages()) {
install.packages("remotes")
}
remotes::install_github("USEPA/rExpertQuery", ref = "develop", dependencies = TRUE, force = TRUE)## Using github PAT from envvar GITHUB_PAT. Use `gitcreds::gitcreds_set()` and unset GITHUB_PAT in .Renviron (or elsewhere) if you want to use the more secure git credential store instead.## Downloading GitHub repo USEPA/rExpertQuery@develop##
## ── R CMD build ─────────────────────────────────────────────────────────────────
## * checking for file ‘/tmp/RtmpVpDgsh/remotes224d77840daf/USEPA-rExpertQuery-5f882fb/DESCRIPTION’ ... OK
## * preparing ‘rExpertQuery’:
## * checking DESCRIPTION meta-information ... OK
## * checking for LF line-endings in source and make files and shell scripts
## * checking for empty or unneeded directories
## Omitted ‘LazyData’ and ‘LazyDataCompression’ from DESCRIPTION
## * building ‘rExpertQuery_0.1.0.tar.gz’## Installing package into '/home/runner/work/_temp/Library'
## (as 'lib' is unspecified)This vignette also requires additional packages which can be installed using the code chunk below:
# # list of additional required packages
# demo.pkgs <- c("datasets", "data.table", "dplyr", "DT", "ggplot2", "httr", "leaflet", "maps", "plotly", "sf", "usdata", "usmap", "stringi")
#
# # install additional packages not yet installed
# installed_packages <- demo.pkgs %in% rownames(installed.packages())
#
# if (any(installed_packages == FALSE)) {
# install.packages(demo.pkgs[!installed_packages])
# }
# load additional packages for this vignette
library(datasets)
library(data.table)
library(dplyr)
library(DT)
library(ggplot2)
library(httr)
library(leaflet)
library(maps)
library(plotly)
library(sf)
library(usdata)
library(usmap)
library(stringi)As a final setup step, we will turn off scientific notation so that any catchment id numbers from our queries are displayed correctly.
# turn off scientific notation (for catchment ids)
options(scipen = 999)API Key
Most of the rExpertQuery functions require an API key. Users can obtain their own by using Expert Query’s API Key Signup Form.
To run this vignette, we can use the following test key. Users should obtain their own API key if they will be using rExpertQuery to develop their own workflows.
# load test api key
testkey <- "53BVce47MQ3KXKibjx35g4ojaDQGh8qWfbdO8cE0"rExpert Query Functions
There are ten exported functions in rExpertQuery. The first eight allow users to query ATTAINS data via Expert Query web services. These are:
- EQ_Actions() - queries and returns ATTAINS actions data
- EQ_ActionsDocuments() - queries and returns ATTAINS Actions Documents, including a keyword search
- EQ_Assessments() - queries and returns ATTAINS Assessments (default is for latest cycle)
- EQ_AssessmentUnits() - queries and returns ATTAINS Assessment Units (default is for Active AUs)
- EQ_AUsMLs() - queries and returns ATTAINS Assessment Units with Monitoring Location data
- EQ_CatchCorr() - queries and returns ATTAINS Catchment Correspondence data (memory intensive)
- EQ_Sources() - queries and returns ATTAINS Sources data
- EQ_TMDLs() - queries and returns ATTAINS TMDL data
These first eight functions are analogous to the data profiles that can be queried through the Expert Query User Interface.
An additional function allows users to download the Expert Query National Extracts of ATTAINS data which are updated weekly. This function should be used when national data are desired or when a query designed in one of the previous eight functions would exceed the Expert Query web services limit of one millions rows. This function is:
- EQ_NationalExtract() - returns national extract for Actions, Assessments, Assessment Units, Assessment Units with Monitoring Locations, Catchment Correspondence, Sources, or TMDLS based on user selection
The tenth function relies on ATTAINS web services to provide allowable domain values for some rExpertQuery parameters:
- EQ_DomainValues() - returns domain values for some rExpertQuery function params
Demo Structure
In this session, we will use rExpertQuery functions, along with some common R packages for data wrangling and visualization to answer some commonly asked questions with Expert Query data. Members of the ATTAINS team will be monitoring the chat for questions. If there are many questions regarding a particular function, we may pause at the end of that function’s material to review. Otherwise, questions are welcome at the end of the demo as time permits or via e-mailing the ATTAINS team at attains@epa.gov.
EQ_NationalExtract
EQ_NationalExtract() provides an efficient method for importing the Expert Query National Extracts. This function requires one argument, “extract” to specify which of the National Extracts to download. The following examples show how to download the Assessments and Assessments with Monitoring Locations extracts. The extracts are all large files and some may not open in R if there is not enough memory available.
-
What is the most commonly reported impairment nationally?
After downloading the national assessments data, we’ll need to filter to include only the latest cycle. Next, we can filter for rows containing parameter causes, and summarize how many times each parameter was listed as a cause of impairment. The additional packages used to answer this question are dplyr (data manipulation) and DT (interface to DataTables library).
# import natioanl assessments profile assessments.nat <- rExpertQuery::EQ_NationalExtract("assessments")## [1] "EQ_NationalExtract: downloading Assessments Profile (Expert Query National Extract). It was last updated on May 31, 2025 at 02:22 AM UTC." ## [1] "EQ_NationalExtract: unzipping Assessments Profile (Expert Query National Extract)." ## [1] "EQ_NationalExtract: opening Assessments Profile (Expert Query National Extract)."# filter for latest assessments assessments.nat <- assessments.nat %>% dplyr::group_by(organizationId) %>% dplyr::slice_max(reportingCycle) %>% dplyr::select(-objectId) %>% dplyr::distinct() %>% dplyr::ungroup() # count impairments count.impair <- assessments.nat %>% # filter to retain only parameter causes dplyr::filter(parameterStatus == "Cause") %>% # subset data to find unique combinations dplyr::select(organizationId, assessmentUnitId, parameterName) %>% # remove duplicates dplyr::distinct() %>% # group by parameterName dplyr::group_by(parameterName) %>% # count listings for each parameterName dplyr::summarise(count = length(assessmentUnitId)) %>% # arrane in descending order dplyr::arrange(desc(count)) # create data table of impairment count results DT::datatable(count.impair, options = list(pageLength = 10, scrollX = TRUE)) -
Which waters are impaired for SELENIUM nationally? Which states contain waters impaired for SELENIUM?
Again, we’ll use the national assessments data filtered to the latest cycle. Then we will filter for rows where the parameter is both selenium and listed as a cause. The additional packages used in this step are dplyr and DT.
# filter assessments data set for param.nat <- assessments.nat %>% # filter for selenium and cause dplyr::filter(parameterName == "SELENIUM" & parameterStatus == "Cause") # data table of selenium impairments DT::datatable(param.nat, options = list(pageLength = 5, scrollX = TRUE))Then we can create a list of states that have at least one selenium impairment listed. We can also visualize this information on a map, with states containing selenium impairments filled in blue. To build this map, we first create a data frame with all 50 states and a “SeleniumImpairment” column populated with “yes” or “no” to indicate whether it has any listed selenium impairments.
In addition to dplyr, this code chunk also uses usdata (US demographic data), datasets (collection of data sets for use in R), usmap (US choropleth plotting) and ggplot2 (creating graphics).
# filter to create df of states that have at least one selenium impairment} state.names.sel <- param.nat %>% # select state column from filtered df dplyr::select(state) %>% # retain only distinct values for state dplyr::distinct() %>% # change state abbreviation to full state name dplyr::mutate(state = usdata::abbr2state(state)) %>% # create a list from the df dplyr::pull() # read in a list of all 50 state names state.names <- datasets::state.name # create df with of all 50 state names sel.map.data <- as.data.frame(state.names) %>% # rename state.name column to state dplyr::rename(state = state.names) %>% # create additional column to indicate if state has recorded selenium impairments ("yes") or not ("no") dplyr::mutate(SeleniumImpairment = factor(ifelse(state %in% state.names.sel, "yes", "no")))# create map, specifying data and values usmap::plot_usmap( regions = "state", data = sel.map.data, values = "SeleniumImpairment", color = "black" ) + # fill states with selenium impairments with blue ggplot2::scale_fill_manual( values = c(`no` = "white", `yes` = "blue") ) + # remove legend ggplot2::theme(legend.position = "none")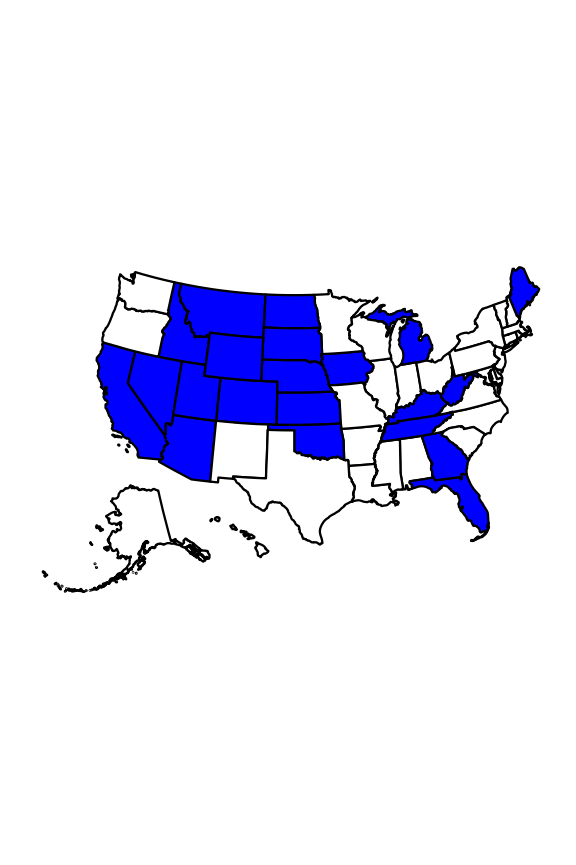
Fig 1. States with selenium listed as a cause of impairment for any waters are shown in blue.
-
Which waters have a TMDL and are fully supporting nationally?
Starting with the filtered data frame of latest cycle assessments, we can filter for Fully Supporting waters with an asssociatedActionType of “TMDL”. Again, we can use a combination of dplyr and DT, for data manipulation and creating a data table, respectively.
# filter national assessments to includ only Fully Supporting w/ TMDL tmdl.fs <- assessments.nat %>% dplyr::filter(overallStatus == "Fully Supporting", associatedActionType == "TMDL" ) %>% # select columns to review dplyr::select(organizationId, state, assessmentUnitId, assessmentUnitName, associatedActionId) %>% # group by state and assessmentUnit dplyr::group_by(state, assessmentUnitId, assessmentUnitName) %>% # create column continaing all actionIds associated with the assessmentUnit dplyr::mutate(associatedActionIds = paste(unique(associatedActionId), collapse = ", ")) %>% # remove original associatedActionId column dplyr::select(-associatedActionId) %>% # retain only distinct rows dplyr::distinct() # fully supporting waters with tmdls data table DT::datatable(tmdl.fs, options = list(pageLength = 5, scrollX = TRUE)) -
What are the impaired waters (cat 4 and 5 nationally)?
The workflow here is similar to the previous question, relying on dplyr and DT. This time we are retaining a few more columns for inclusion in the data table and are filtering by epaIRCategory for category 4 and 5 waters.
# select subset of columns to include imp.waters <- assessments.nat %>% dplyr::select(organizationId, organizationName, organizationType, assessmentUnitId, assessmentUnitName, assessmentDate, epaIrCategory, waterType, waterSize, waterSizeUnits) %>% # retain only distinct rows dplyr::distinct() %>% # filter for category 4 and 5 waters dplyr::filter(epaIrCategory %in% c("4", "5"))The resulting data frame is large, with 128,870 unique impaired waters nationally. Due to its size, we’ll include a random subset of 250 results in a data table to review during the demo. You can view the full results by viewing the imp.waters df.
# data table with random 250 impaired waters DT::datatable(imp.waters %>% dplyr::slice_sample(n = 250), options = list(pageLength = 5, scrollX = TRUE)) -
Which states have included any information about monitoringLocations in ATTAINS? Which states have included any monitoringLocationDataLinks?
To answer this question, we’ll need to import a new national extract. As we are curious about monitoringLocations and monitoringLocationDataLinks, we will want to start with the Assessment Units with Monitoring Locations data set. The param value for this in EQ_NationalExtract is “au_mls”. Additional packages needed are dplyr and DT. We’ll use some additional options in DT to highlight useful rows in the final data table.
# import national extract for assessment units/monitoring locations nat.ausmls <- rExpertQuery::EQ_NationalExtract("au_mls")The next step is to filter the data frame and retain only rows that contain a value for monitoringLocationId and are relevant to states. The resulting data frame, called “filtered.mls” will allow us to determine (1) which organizations have included monitoringLocationIdentifiers and (2) which organizations have included monitoringLocationDataLinks.
To find the organizations with monitoringLocationIdentifiers, we will select only the rows organizationId, organizationName, and organizationType and retain the unique rows.
To find the organizations with monitoringLocationIdentifiers, we will apply an additional filter and keep only rows that have a value in the monitoringLocationDataLink column. Then select only the rows organizationId, organizationName, and organizationType and retain the unique rows.
# filter for rows that contain monitoringLocationIds and are from State orgs filtered.mls <- nat.ausmls %>% dplyr::filter(monitoringLocationId != "", !is.na(monitoringLocationId), organizationType == "State") # start with data set filtered to include only rows with MonitoringLocationIds orgs.mls <- filtered.mls %>% # select columns to describe orgs dplyr::select(organizationId, organizationName, organizationType) %>% dplyr::distinct() # start with data set filtered to include only rows with MonitoringLocationIds links.mls <- filtered.mls %>% # filter for orgs with monitoringLocationDataLinks dplyr::filter(monitoringLocationDataLink != "", !is.na(monitoringLocationDataLink)) %>% # select columns to describe orgs dplyr::select(organizationId, organizationName, organizationType) %>% # retain distinct rows dplyr::distinct()There are 21 with monitoringLocationId data in ATTAINS. Of these states, 3 also contain monitoringLocationDataLinks. We can create a data table to display this information and apply some conditional formatting to highlight the states with monitoringLocationIdentifiers in blue and those with monitoringLocationIdentifiers and monitoringLocationDataLinks in orange.
# create data frame summarizing which states have monitoringLocationIds and monitoringLocationDataLinks in ATTAINS ml.table <- nat.ausmls %>% # create df of all orgs dplyr::select(organizationId, organizationName, organizationType) %>% # retain only distinct rows dplyr::distinct() %>% # add columns indicating whether the org has any data in ATTAINS for monitoringLocationId and monitoringLocationDataLink dplyr::mutate(monitoringLocationId = ifelse(organizationId %in% orgs.mls$organizationId, "yes", "no"), monitoringLocationDataLink = ifelse(organizationId %in% links.mls$organizationId, "yes", "no")) # create data table DT::datatable(ml.table, options = list(pageLength = 10, scrollX = TRUE)) %>% # highlight rows for orgs that have monitoringLocationId in blue DT::formatStyle( "monitoringLocationId", target = "row", backgroundColor = styleEqual(c("yes"), c("#66c3c5"))) %>% # highlight rows for orgs that also have data for data links in organge DT::formatStyle( "monitoringLocationDataLink", target = "row", backgroundColor = styleEqual(c("yes"), c("#f9b97d")))
EQ_Assessments
EQ_Assessments()queries ATTAINS Assessments data. For many use cases, querying Assessments for a single organization or state is desirable. The next example shows how to query by a single state for the latest cycle. The default for the function is to query for the latest cycle. If you would like to search for an older cycle, you can set the “report_cycle” parameter equal to the desired cycle year in YYYY format.
-
How many stream miles in Kansas are impaired for primary contact recreation in the latest cycle?
First, we can construct our query to return only Primary Contact Recreation Assessments from the latest cycle from Kansas for streams. The remaining steps can be accomplished with dplyr functions to retain unique combinations of assessmentUnitId, assessmentUnitName, waterSize and waterSizeUnits, then summing the waterSize column to find the number of stream miles.
# set up query to search assessments for KS, primary contact recreation use, and stream, default behavior is to query latest report cycle ks.imp.streams <- rExpertQuery::EQ_Assessments(statecode = "KS", use_name = "Primary Contact Recreation", api_key = testkey, water_type = "STREAM") # calculate total water size ks.miles <- ks.imp.streams %>% # select required columns dplyr::select(assessmentUnitId, assessmentUnitName, waterSize, waterSizeUnits) %>% # retain distinct rows dplyr::distinct() %>% # sum water size dplyr::summarize(totalWaterSize = sum(waterSize)) %>% dplyr::pull()There are 11,129.78 stream miles in Kansas impaired for primary contract recreation use.
-
What waters in Montana are impaired due to Zinc? How many different uses are zinc impairments associated with in Montana?
We start by querying for Montana assessments with zinc as a cause and can use dplyr and plotly (graphing library) to summarize the results by useName and create a bar plot of the results.
# query MT's latest assessments for zinc as a cause MT.impair <- rExpertQuery::EQ_Assessments(statecode = "MT", api_key = testkey, param_name = "ZINC", param_status = "Cause" )## [1] "EQ_Assessments: The current query will return 123 rows."Next we summarize the uses associated with zinc impairments and count the number of unique assessmentUnitIdentifiers for each use to facilitate creating a bar plot.
# summarize by useName MT.impair.use <- MT.impair %>% dplyr::group_by(useName) %>% # count AUIDs by use dplyr::summarize(count = length(unique(assessmentUnitId)))There are 3 uses with zinc as a cause in Montana: Agricultural, Aquatic Life, and Drinking Water . As our last step, we’ll create the bar plot.
plotly::plot_ly(data = MT.impair.use, x = ~useName, y = ~count, type = "bar" )Fig 2. Zinc impairments by useName for Montana’s latest assessment cycle.
-
Which waters are new to category 5 in the 2024 cycle for Illinois? Which categories did they move from?
This is another question that is well suited to using dplyr and plotly in addition to EQ_Assessments. The query is for category 5 waters from Illinois from the latest reporting cycle. In order to compare reporting cycles, we’ll also need to provide a value for the previous cycle and run the query again, this time for the previous reporting cycle and without specifying the category.
# get cat 5 waters from 2024 assessment cycle IL.latest <- rExpertQuery::EQ_Assessments(statecode = "IL", epa_ir_cat = "5", api_key = testkey) # select required columns from latest cycle data IL.latest <- IL.latest %>% dplyr::select(assessmentUnitId, epaIrCategory) %>% # retain unique rows dplyr::distinct() # get all assessments from previous cycle IL.previous <- rExpertQuery::EQ_Assessments(statecode = "IL", report_cycle = 2022, api_key = testkey)Next we’ll need to filter the previous cycle results to retain only the assessment units that are category 5 waters in the latest cycle. This allows us to combine the latest and previous cycle data frames and filter them to create a data frame which contains only the assessment units that changed from some other category to category 5 from the previous cycle to the latest cycle.
# filter previous cycle to include only cat 5 waters from 2024 IL.previous <- IL.previous %>% # retain only cat 5 waters from latest cycle dplyr::filter(assessmentUnitId %in% IL.latest$assessmentUnitId) %>% # select auid and category dplyr::select(assessmentUnitId, epaIrCategory) %>% # rename to indicate previous cycle dplyr::rename(epaIrCategory.previous = epaIrCategory) # combine latest and previous cycle dfs IL.combine <- IL.latest %>% dplyr::left_join(IL.previous, dplyr::join_by(assessmentUnitId)) # create df of the auids that changed to cat 5 in 2024 IL.changes <- IL.combine %>% dplyr::filter(epaIrCategory != epaIrCategory.previous) %>% dplyr::distinct()Finally, we can summarize by counting the categories the new category 5s were previously in and create a pie chart with a default color scheme to visualize.
# summarize what cats the new cat 5s were in previously IL.change.sum <- IL.changes %>% dplyr::group_by(epaIrCategory.previous) %>% dplyr::summarise(count = dplyr::n_distinct(assessmentUnitId))# create plotly plot plotly::plot_ly() %>% # add pipe based IL.change.sum plotly::add_pie(data = IL.change.sum, ~count, labels = ~epaIrCategory.previous, marker = list( line = list(color = "white", width = 2) ), type = "pie", hoverinfo = "value", outsidetextfont = list(color = "black"), sort = FALSE ) %>% # add legend plotly::layout(legend = list(orientation = "h", xanchor = "center", x = 0.5))Fig 3. New category 5 waters in Illinois in the 2024 cycle, summarized by their 2022 categories.
EQ_AssessmentUnits
EQ_AssessmentUnits() queries ATTAINS Assessment Units data. The default is to only return “Active” assessment units, but input params can be adjusted to query for “Retired” or “Historical” assessment units in addition to or instead of “Active”.
-
What are the retired assessment units for Florida?
This can be answered with a simple EQ_AssessmentUnits query specifying Florida with an assesment unit status of “Retired” without using functions from any additional packages. For easier review, we’ll create a datatable with DT.
# search for retired assessment units in Florida FL.ret.auids <- rExpertQuery::EQ_AssessmentUnits(statecode = "FL", au_status = "Retired", api_key = testkey)## [1] "EQ_AssessmentUnits: The current query will return 35 rows."
EQ_TMDLs
EQ_TMDLs() queries ATTAINS TMDL data and can utilize many search parameters.
-
What are all the TMDLs in HI? How many are there (counted as unique combinations of actionID/pollutant/assessmentUnitId)? How many are associated with each pollutantGroup?
To answer this question we’ll use dplyr and plotly. The EQ_TMDLs query is simple, specifying only the state of Hawaii since we want to return all of Hawaii’s TMDLs.
# query for Hawaii TMDLs HI.tmdls <- rExpertQuery::EQ_TMDLs(statecode = "HI", api_key = testkey)The 536 rows can be reviewed in the data frame. Keep in mind that if we want to count one TMDL as one unique combination of assessmentUnitIdentifier, pollutant and actionId, we can’t simply count the number of rows as as each assessmentUnitIdentifier/pollutant/actionId may be associated with multiple parameters, meaning it will be listed more than once. The code chunk below demonstrates one way to determine the number of TMDLs by unique assessmentUnitIdentifier/pollutant/actionId. We can than visualize TMDLs related to each pollutantGroup with a simple bar graph.
# determine the number of tmdls HI.count <- HI.tmdls %>% # select required columns dplyr::select(assessmentUnitId, assessmentUnitName, actionId, actionName, pollutant, pollutantGroup, fiscalYearEstablished, actionAgency, planSummaryLink) %>% # retain unique rows dplyr::distinct() # determine number per group HI.sum <- HI.count %>% # groupy by pollutantGroup dplyr::group_by(pollutantGroup,) %>% # count unique auids per pollutantGroup dplyr::summarize(count = dplyr::n_distinct(assessmentUnitId)) %>% # if pollutantGroup is missing, assign to group named "NULL" dplyr::mutate(pollutantGroup = ifelse(is.na(pollutantGroup), "NULL", pollutantGroup))# create bar plot plotly::plot_ly( data = HI.sum, x = ~pollutantGroup, y = ~count, type = "bar" )Fig 4. Number of TMDLs per pollutantGroup in Hawaii.
There are 168 TMDLs in Hawaii.
-
How many TMDL projects (by actionId) were approved so far in FY 2025 in R10?
This question can be answered simply with a well crafted query and a single dplyr function. The 2025 fiscal year began on October 1, 2024, so that will be set as the tmdl_start_date parameter along with specifying 10 for the region.
# r10 query for FY 2025 TMDLs tmdl.proj <- rExpertQuery::EQ_TMDLs(region = 10, tmdl_date_start = "2024-10-01", api_key = testkey) # count distinct actionIds tmdl.proj.count <- dplyr::n_distinct(tmdl.proj$actionId)There are TMDL projects (by actionID) approved so far in FY 2025. In order to create a user-friendly DT datatable, we will have to take a few more steps. The columns, assessmentUnitId, pollutant, and addressedParameter are likely to have more than one value per actionId. In order to create a readable table with only one row per actionId, we will use dplyr functions to manipulate the data frame so that for example, multiple pollutants will be listed together in the same row.
tmdl.sum <- tmdl.proj %>% # select columns dplyr::select(region, state, organizationId, organizationName, organizationId, actionId, actionName, assessmentUnitId, pollutant, addressedParameter, tmdlDate, planSummaryLink) %>% # retain unique rows dplyr::distinct() %>% # group by actionId dplyr::group_by(actionId) %>% # create new columns to address issues with mulitple rows dplyr::mutate(assessmentUnitIds = paste(sort(unique(assessmentUnitId)), collapse = ", " ), pollutants = paste(sort(unique(pollutant)), collapse = ", "), addressedParameters = paste(sort(unique(addressedParameter)), collapse = ", ")) %>% dplyr::select(-assessmentUnitId, -pollutant, -addressedParameter) %>% # retain unique rows dplyr::distinct() # create data table DT::datatable(tmdl.sum %>% rExpertQuery::EQ_FormatPlanLinks(), escape = FALSE, options = list(pageLength = 2, scrollX = TRUE))
EQ_Actions
EQ_Actions()queries ATTAINS Actions data by a variety of user supplied parameters.
-
Which actions in Delaware should be included in measures? How many actions (by actionIds) are in this group?
We can use dplyr and DT with the query results to find the answer and display the relevant actionIds. For the query, we use EQ_Actions to search for Deleware actions with the “in_meas” parameter set to “Yes”. Then we count the distinct actionIds.
# query Actions from Delaware that are included in Measures DE.meas <- rExpertQuery::EQ_Actions(statecode = "DE", in_meas = "Yes", api_key = testkey)## [1] "EQ_Actions: The current query will return 649 rows."# count unique actionIds DE.count <- dplyr::n_distinct(DE.meas$actionId)There are 480 actionIds that should be included when calculating measures for Delaware. In a similar process to the final TMDL example, we can create a data table with one row per actionId.
# determine unique actionIds DE.actids <- DE.meas %>% # select columns dplyr::select(actionType, actionId, actionName, actionAgency, parameter, completionDate, assessmentUnitId, fiscalYearEstablished, planSummaryLink) %>% # retain unique rows dplyr::distinct() %>% # group by actionId dplyr::group_by(actionId) %>% # create new columns to address issues with mulitple rows dplyr::mutate(parameters = paste(unique(parameter), collapse = ", "), assessmentUnitIds = paste(unique(assessmentUnitId), collapse = ", ")) %>% dplyr::select(-parameter, -assessmentUnitId) %>% # retain unique rows dplyr::distinct()DT::datatable(DE.actids %>% rExpertQuery::EQ_FormatPlanLinks(), escape = FALSE, options = list(pageLength = 5, scrollX = TRUE)) -
How many actions (by actionId) were established in R3 in FY 2024? What was the most common type of action established in R3 in FY 2024?
For this query, we will set region equal to 3 and both fisc_year_start and fisc_year_end to 2024. Then use dplyr and DT to summarize and display results as in the first EQ_Actions example.
# query for R3 FY2024 actions act.fy.24 <- rExpertQuery::EQ_Actions(region = 3, fisc_year_start = 2024, fisc_year_end = 2024, api_key = testkey) # count unique actionIds n_act.fy.24 <- dplyr::n_distinct(act.fy.24$actionId)There were 7 unique actions by actionId established in R3 in FY 2024. We can create data table with counts for each actionType.
act.fy.24.counts <- act.fy.24 %>% # select columns dplyr::select(state, organizationId, organizationName, actionId, actionName, actionType) %>% # retain unique rows dplyr::distinct() %>% # group by action type dplyr::group_by(actionType) %>% # count actionId per each action type dplyr::summarize(count = length(unique(actionId))) DT::datatable(act.fy.24.counts)
EQ_ActionsDocuments
EQ_ActionsDocuments() queries ATTAINS Actions Documents data with user supplied parameters. One of its unique features is the ability to search the documents themselves by keyword or phrase. The results are ranked by percentage match, with higher percentages indicating a better match with the keyword.
-
What are all the actions documents that contain the keyword “nutrient”?
The only query parameter we will provide is the keyword “nutrient”.
# Actions Documents containing "nutrient" nutrient.doc <- rExpertQuery::EQ_ActionsDocuments(doc_query = "nutrient", api_key = testkey )This query yields 17,659 results. As well as providing the actionId and documentName for all results, EQ_Actions() also returns the column “actionDocumentUrl” containing the URL to link to the document. Because the query yields so many results, we’ll take a look at the highest ranking 50 matches in a DT data table to understand the structure of the results.
-
How many actions documents that contain the keyword “nutrient” are from each region?
To answer this question, we can count the number of unique actionIds per region from the initial “nutrients” keyword query. We can display the results in a plotly bar plot.
nutrient.doc.regions <- nutrient.doc %>% # group by region dplyr::group_by(region) %>% # count action ids by region dplyr::summarize(count = length(unique(actionId)))# create bar plot plotly::plot_ly(data = nutrient.doc.regions, x = ~region, y = ~count, type = "bar" )Fig 5. Number of documents containing the keyword ‘nutrient’ by EPA Region.
EQ_AUsMLs
-
Which monitoringLocations correspond to assessmentUnits in Alaska? How many monitoringLocations are associated with assessmentUnits? Which organizations are the monitoringLocations from?
We start by querying for all monitoringLocations in Alaska and then use dplyr functions to retain only rows that have a value for monitoringLocation. We can create a DT data table to review the results. We can also use the base R functions sort and unique to make a list of the organizations represented in the monitoringLocationOrgId column and organize them alphabetically.
# query for assessment units and monitoring locations from Alaska ak.mls <- rExpertQuery::EQ_AUsMLs(statecode = "AK", api_key = testkey) # filter for assessment units with monitoring locations from Alaska filt.ak.mls <- ak.mls %>% # filter for rows with a valule for monitoring location id dplyr::filter(!is.na(monitoringLocationId), monitoringLocationId != "") %>% # select rows for review dplyr::select(assessmentUnitId, assessmentUnitName, monitoringLocationId, monitoringLocationOrgId) %>% # retain unique rows dplyr::distinct() # list of orgs inlcuded in monitoringLocationOrgId ak.orgs <- sort(unique(filt.ak.mls$monitoringLocationOrgId)) DT::datatable(filt.ak.mls, options = list(pageLength = 10, scrollX = TRUE))There are 208 monitoringLocationIds associated with assessmentUnits in Alaska. The monitoringLocationIds come from 5 different organizations: `ADOT&PF, AKDECWQ, Kenai Watershed Forum, National Park Service, and USFWS_ALASKA .
-
Are there active assessmentUnits without any listed monitoringLocations in Alaska?
This question can be answered by filtering the initial Alaska query results for assessmentUnitIds without any corresponding monitoringLocationIdentifiers.
# filter Alaska data set for assessmentUnitIds without listed monitoringLocationIds no.ml.ak <- ak.mls %>% dplyr::filter(!assessmentUnitId %in% filt.ak.mls$assessmentUnitId) # count number of unique asessment units without listed monitoringLocationIds n_no.ml.ak <- dplyr::n_distinct(no.ml.ak$assessmentUnitId)There are 636 active assessmentUnits in Alaska without monitoringLocations listed in ATTAINS.
EQ_Sources
-
Which states in Region 6 have listed probable Sources in their latest report cycle?
Start by querying for EQ_Source for region 6. Then use dplyr and usdata to manipulate the data and create a list of the region 6 states that list probable sources for any assessments in their latest report cycle.
# query for probable sources in the latest assessment cycle in R6 r6.sources <- rExpertQuery::EQ_Sources(region = 6, api_key = testkey) # filter for states with listed probable sources r6.sources.filter <- r6.sources %>% # remove all rows without a listed source dplyr::filter(!is.na(sourceName)) %>% # select state dplyr::select(state) %>% # retain distinct state dplyr::distinct() %>% # create list dplyr::pull() %>% # convert abbreviation to state name usdata::abbr2state() %>% # sort alphabetically sort()Arkansas, Louisiana, New Mexico, Oklahoma and Texas all listed sources in their latest report cycles.
-
What are the most commonly listed sources in the latest report cycle in each Region 6 state?
Using the same initial query results from the previous question, we can use dplyr functions to count the number of times a probable source is listed by each state, select the most one from each state and make a a DT data table of the results.
# count the number of times a probable source is listed for each state r6.sources.summarize <- r6.sources %>% dplyr::select(state, organizationId, assessmentUnitId, sourceName) %>% dplyr::distinct() %>% dplyr::group_by(state, organizationId, sourceName) %>% dplyr::summarize(count = length(assessmentUnitId))%>% dplyr::ungroup() %>% dplyr::group_by(state) %>% dplyr::slice_max(count) # data table for max count DT::datatable(r6.sources.summarize)
EQ_CatchCorr
EQ_CatchCorr() queries ATTAINS Catchment Correspondance data. These queries return large files, particularly if a large geospatial area is defined, so best practice is to make your query as specific as possible. Many machines may not have enough memory to load an entire state’s worth of catchment correspondence data in an R session. It is also possible to focus on a specific assessment unit in catchment correspondence queries.
-
Which catchments correspond to assessmentUnitId “IL_N-99” ?
We can start with a very focused query to retrieve information for the Illinios assessment unit “IL_N-99”. Then we’ll use dplyr functions to retain the unique rows make a a DT data table.
# query for IL_N-99 catchment data il.n99.catch <- rExpertQuery::EQ_CatchCorr(statecode = "IL", auid = "IL_N-99", api_key = testkey) il.n99.catch <- il.n99.catch %>% # remove objectId dplyr::select(-objectId) %>% # retain unique rows dplyr::distinct() # create data table DT::datatable(il.n99.catch, options = list(pageLength = 10, scrollX = TRUE)) -
How large of a catchment area corresponds to “IL_N-99”?
We will need some additional information in order to answer this question. We could get catchment areas from the nhdplusTools package or from ATTAINS geospatial web services. For this example, we will use ATTAINS geospatial web services as this will also allow us to create a map of the catchments and assessment unit. This example utlizes a few new R packages, httr (wrapper for curl packge), leaflet (interactive maps), and sf (spatial vector data).
The first step is to set up query parameters for ATTAINS geospatial webservices. Then query the assessment lines and catchment associations layers to retrieve the geospatial data for the assessment unit and its associated catchments. The data from the catchment association layer allows us to sum the total area for all the catchments associated with IL_N-99.
# set up query params for ATTAINS geospatial web services query.params <- list( where = "assessmentunitidentifier IN ('IL_N-99')", outFields = "*", f = "geojson" ) # query ATTAINS geospatial assessments lines layer lines.response <- httr::GET("https://gispub.epa.gov/arcgis/rest/services/OW/ATTAINS_Assessment/MapServer/1/query?", query = query.params) # read content from ATTAINS geospatial assessments lines layer lines.geojson <- httr::content(lines.response, as = "text", encoding = "UTF-8") # get sf from ATTAINS geospatial assessments lines layer lines.sf <- sf::st_read(lines.geojson, quiet = TRUE) # query ATTAINS geospatial catchment association layer catch.response <- httr::GET("https://gispub.epa.gov/arcgis/rest/services/OW/ATTAINS_Assessment/MapServer/3/query?", query = query.params) # read content from ATTAINS geospatial catchment association layer catch.geojson <- httr::content(catch.response, as = "text", encoding = "UTF-8") # # get sf from ATTAINS geospatial assessments lines layer catch.sf <- sf::st_read(catch.geojson, quiet = TRUE) # sum the catchment areas that correspond to IL_N-99 sum.catch <- catch.sf %>% dplyr::summarise(total = sum(areasqkm)) %>% sf::st_drop_geometry() %>% dplyr::pull()The total catchment area associated with IL_N-99 is 35.6019.
To create map of IL_N-99 and and its associated catchments, we can create a leaflet map with the default Open Street Map tile then add the catchment polygons with a popup contain the nhdplusid and area in sq km. The final step is to add the assessment unit line.
# create leaflet map leaflet::leaflet() %>% # add open street map layer leaflet::addTiles() %>% # add catchments, include popups with nhdplus id and area in sq km leaflet::addPolygons(data = catch.sf, color = "darkorange", popup = paste0(catch.sf$nhdplusid, "<br>", catch.sf$areasqkm, " sq km")) %>% # added assessment id leaflet::addPolylines(data = lines.sf, color = "blue")Fig. 6 Map of assessment unit IL_N-99 (blue) and its associated catchments (orange).
EQ_DomainValues
EQ_DomainValues is unique in the rExpertQuery packages as it relies on ATTAINS web services, rather than Expert Query web services in order to provide allowable domain values. It was created to provide guidance for users on allowable values for queries. The function returns all information available from ATTAINS web services about the selected domain and prints a message informing the user which column contains the value that should be used in queries.
-
Which parameters can EQ_DomainValues provide values for?
Running the EQ_DomainValues function with no value provided for the “domain” argument will return a list of the parameter names that it can return domain values for.
# run EQ_DomainValues with no value provided for "domain" to return list of params domain.vals <- EQ_DomainValues()## [1] "EQ_DomainValues: getting list of available domain names."EQ_DomainValues can provide values for the following query parameters: act_agency, act_status, act_type, ad_param, assess_basis, assess_methods, assess_types, au_status, cause, delist_reason, doc_type, file_type, loc_type, org_id, org_name, param_attain, param_group, param_name, param_state_ir_cat, param_status, source_scale, source_type, statecode, use_name, use_support and water_type .
-
How many different values for “water_type” can be used in rExpertQuery functions?
To find the allowable values for “water_type” in rExpertQuery function, we can run EQ_DomainValues again, this time specifying “water_type”.
water.types <- rExpertQuery::EQ_DomainValues("water_type")## [1] "EQ_DomainValues: For water_type the values in the name column of the function output are the allowable values for rExpert Query functions."The printed message informs us that the “name” column contains the values for use in water_type queries for rExpertQuery functions. For water_type there are 66 unique values. They are: BAY; BAYOU; BEACH; BLACKWATER SYSTEM; CHANNEL; CIRQUE LAKE; COASTAL; COASTAL & BAY SHORELINE; CONNECTING CHANNEL; CREEK; CREEK, INTERMITTENT; DITCH OR CANAL; DRAIN; ESTUARY; ESTUARY, FRESHWATER; FLOWAGE; GREAT LAKES BAYS AND HARBORS; GREAT LAKES BEACH; GREAT LAKES CONNECTING CHANNEL; GREAT LAKES OPEN WATER; GREAT LAKES SHORELINE; GULCH; GULF; HARBOR; IMPOUNDMENT; INLAND LAKE SHORELINE; INLET; ISLAND COASTAL WATERS; LAGOON; LAKE; LAKE, FRESHWATER; LAKE, NATURAL; LAKE, PLAYA; LAKE, SALINE; LAKE, SPRINGS; LAKE, WILD RICE; LAKE/RESERVOIR/POND; MARSH; OCEAN; OCEAN/NEAR COASTAL; POND; RESERVOIR; RESERVOIR EMBAYMENT; RIVER; RIVER, TIDAL; RIVER, WILD RICE; RIVERINE BACKWATER; SINK HOLE; SOUND; SPRING; SPRINGSHED; STREAM; STREAM, COASTAL; STREAM, EPHEMERAL; STREAM, INTERMITTENT; STREAM, PERENNIAL; STREAM, TIDAL; STREAM/CREEK/RIVER; WASH; WATERSHED; WETLAND; WETLANDS, DEPRESSIONAL; WETLANDS, FRESHWATER; WETLANDS, RIVERINE; WETLANDS, SLOPE; and WETLANDS, TIDAL.