(9) A Physiologically Based Pharmacokinetic Model
Dustin Kapraun
2026-03-01
Source:vignettes/pbpk_demo.Rmd
pbpk_demo.RmdModel Description
Like any pharmacokinetic (PK) model, a physiologically based pharmacokinetic (PBPK) model is a mathematical description of the absorption, distribution, metabolism, and excretion of a substance by an organism. PBPK models differ from classical PK models in that many of their parameters describe actual anatomical and physiological properties of the organism, such as volumes of and blood flow rates to various organs and tissues. Figure 1 shows a schematic representation of a relatively simple PBPK model for “Substance X.”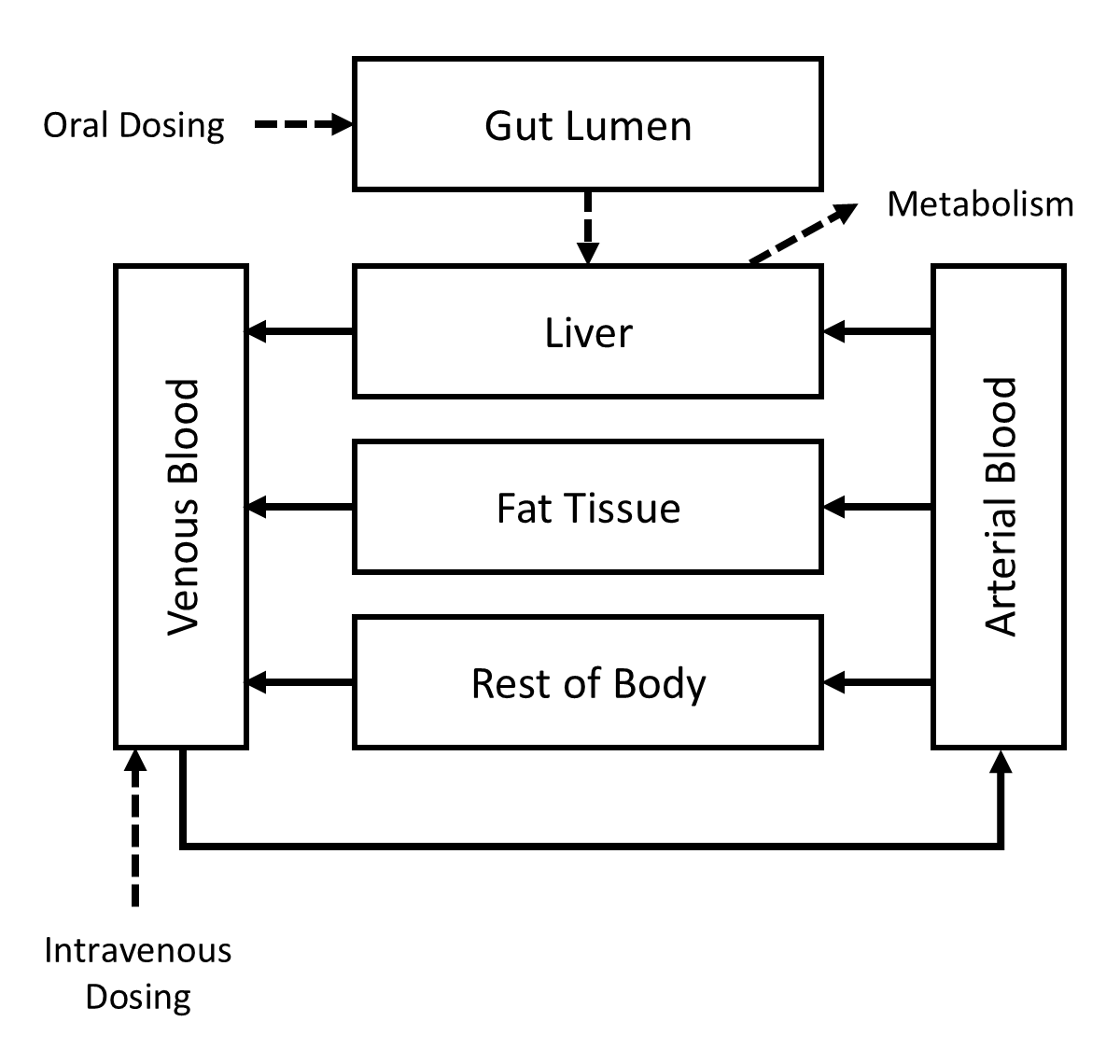
Figure 1: A simple PBPK model
Substance X can enter the body through oral and/or intravenous routes and it is eliminated via saturable metabolism in the liver. The kinetics of distribution are assumed to be “perfusion limited,” so concentrations in different tissues depend on steady state concentration ratios, or “partition coefficients,” for each type of tissue.
The state variables for the PBPK model include:
- , the amount (of Substance X) in the gut lumen (mg);
- , the amount in the liver (mg);
- , the amount in the fat (mg);
- , the amount in the arterial blood (mg);
- , the amount in the venous blood (mg);
- , the amount in the rest of body (mg);
- , the cumulative amount that has been dosed (mg); and
- , the cumulative amount that has been metabolized (mg).
The anatomical and physiological parameters are:
- , the body mass of the organism (kg);
- , the cardiac output constant (L/h/kg, i.e., liters per hour per kg of body mass to the 3/4 power);
- , the constant fractional blood flow to the liver;
- , the constant fractional blood flow to the fat;
- , the constant volume fraction of body mass that is the liver (L/kg);
- , the constant volume fraction of body mass that is the fat (L/kg);
- , the constant volume fraction of body mass that is the arterial blood (L/kg); and
- , the constant volume fraction of body mass that is the venous blood (L/kg).
Values for anatomical and physiological parameters for rats and humans are shown in the following table.
| Parameter | Units | Rat Value | Human Value |
|---|---|---|---|
| kg | 0.25 | 80 | |
| L/h/kg | 15 | 15 | |
| – | 0.21 | 0.26 | |
| – | 0.06 | 0.05 | |
| L/kg | 0.04 | 0.02 | |
| L/kg | 0.07 | 0.21 | |
| L/kg | 0.02 | 0.02 | |
| L/kg | 0.05 | 0.05 |
The chemical-specific parameters for Substance X are:
- , the liver-to-blood partition coefficient;
- , the fat-to-blood partition coefficient;
- , the rest-of-body-to-blood partition coefficient;
- , the maximum metabolic rate constant (mg/h/kg);
- , the affinity constant for saturable metabolism (mg/L); and
- , the first-order oral absorption rate constant (h).
In general, chemical-specific parameters can have different values for different animal species but, in this case, we assume that they have the same values for rats and humans.
The dosing parameters are:
- , the body mass normalized daily oral dose rate (mg/kg/d); and
- , the body mass normalized daily intravenous dose rate (mg/kg/d).
Note that the dosing rates are given in units of mg of Substance X per kg of body mass per day.
To construct the state equations for the model, the following “calculated parameters” are needed:
- , cardiac output (L/h);
- , blood flow rate to the liver (L/h);
- , blood flow rate to the fat (L/h);
- , constant fractional blood flow to the rest of body;
- , blood flow rate to the rest of body (L/h);
- , volume of the liver (L);
- , volume of the fat (L);
- , volume of the arterial blood (L);
- , volume of the venous blood (L);
- , constant volume fraction of body mass that is the rest of body (L/kg);
- , volume of the rest of body (L);
- , maximum rate of metabolism (mg/h);
- , the oral dose rate (mg/h); and
- , the intravenous dose rate (mg/h);
In addition to the model parameters, the following time-varying quantities are needed to construct the state equations:
- , the concentration (of Substance X) in the liver (mg/L) at time ;
- , the concentration in the fat (mg/L) at time ;
- , the concentration in the arterial blood (mg/L) at time ;
- , the concentration in the venous blood (mg/L) at time ;
- , the concentration in the rest of body (mg/L) at time ;
- , the rate of metabolism in the liver (mg/h) at time ; and
- , the rate of absorption from the gut lumen to the liver (mg/h) at time .
The state equations for the PBPK model are:
- ;
- ;
- ;
- ;
- ;
- ;
- ; and
- .
In order to solve an initial value problem for the PBPK model, one needs to provide the values of the basic parameters (but not the calculated parameters) and initial values for all the state variables.
MCSim Model Specification
We used the GNU
MCSim model specification language to implement the simple PBPK
model for Substance X. The complete MCSim model specification file for
this model, pbpk_simple.model, can be found in the
extdata subdirectory of the MCSimMod
package installation directory.
In addition to the state variables that have already been described, we included a state variable that represents the area under the venous blood concentration vs. time curve (AUC), as this is a quantity that is often computed in pharmacokinetic analyses. The AUC for the period from time to time is given by and thus the state equation for this quantity is
Also, in addition to the time-varying quantities previously mentioned, the model includes another output variable, that represents the total amount of substance that has been “lost” vs. time. This amount is calculated as [total (cumulative) amount that has entered in the organism] - [total amount currently in the organism] - [total (cumulative) amount that has been eliminated from the organism]. Therefore, if the model “conserves mass” as it should, the value of should be zero (or very close to zero for numerical solutions to an IVP) at all times .
Note that the model specification file uses text symbols to represent
the state variables, output variables, and parameters in the model. For
example, the state variable
is represented by the text symbol A_L, the output variable
is represented by the text symbol C_L, and the parameter
is represented by the text symbol Q_LC.
Building the Model
First, we load the MCSimMod package as follows.
Using the following commands, we create a model object (i.e., an
instance of the Model class) using the model specification
file pbpk_simple.model that is included in the
MCSimMod package.
# Get the full name of the package directory that contains the example MCSim
# model specification file.
mod_path <- file.path(system.file(package = "MCSimMod"), "extdata")
# Create a model object using the example MCSim model specification file
# "pbpk_simple.model" included in the MCSimMod package.
pbpk_mod_name <- file.path(mod_path, "pbpk_simple")
pbpk_mod <- createModel(pbpk_mod_name)This last command creates a Model object called
pbpk_mod. Once that object is created, we can “load” the
model (so that it’s ready for use in a given R session) as follows.
# Load the model.
pbpk_mod$loadModel()Verifying Conservation of Mass
We can demonstrate that the PBPK model for Substance X conserves mass by performing a simulation in which a rat is orally administered 1 mg/kg/d of Substance X and plotting total amount of substance lost, , vs. time, .
The default values of the anatomical and physiological model
parameters that are provided in the model specification file,
pbpk_simple.model, are for a rat, and these are the values
that are assigned to the parameters when one calls the
loadModel() method. This can be verified by examining the
attribute parms of the Model object
pbpk_mod as follows.
pbpk_mod$parms
#> M Q_CC Q_LC Q_FC V_LC V_FC V_ABC
#> 0.2500000 15.0000000 0.2100000 0.0600000 0.0400000 0.0700000 0.0200000
#> V_VBC P_L P_F P_R V_maxC K_m k_abs
#> 0.0500000 3.5000000 86.5000000 2.3000000 10.0000000 6.0000000 1.2500000
#> D_oral D_IV Q_C Q_L Q_F Q_RC Q_R
#> 0.0000000 0.0000000 5.3033009 1.1136932 0.3181981 0.7300000 3.8714096
#> V_L V_F V_AB V_VB V_RC V_R V_max
#> 0.0100000 0.0175000 0.0050000 0.0125000 0.8200000 0.2050000 3.5355339
#> R_oral R_IV
#> 0.0000000 0.0000000We want to perform a simulation in which a rat is orally administered 1 mg/kg/d of Substance X, so we will change the value of the parameter from 0 to 1.
pbpk_mod$updateParms(c(D_oral = 1))Now, if we examine the values of the model parameters, we can see
that the body mass normalized oral administration rate (represented by
and D_oral) has been updated from 0 to 1 mg/kg/d and the
actual oral administration rate (represented by (represented by
and R_oral), which is a calculated parameter, has also been
updated (from 0 to approximately 0.0104 mg/h).
pbpk_mod$parms
#> M Q_CC Q_LC Q_FC V_LC V_FC
#> 0.25000000 15.00000000 0.21000000 0.06000000 0.04000000 0.07000000
#> V_ABC V_VBC P_L P_F P_R V_maxC
#> 0.02000000 0.05000000 3.50000000 86.50000000 2.30000000 10.00000000
#> K_m k_abs D_oral D_IV Q_C Q_L
#> 6.00000000 1.25000000 1.00000000 0.00000000 5.30330086 1.11369318
#> Q_F Q_RC Q_R V_L V_F V_AB
#> 0.31819805 0.73000000 3.87140963 0.01000000 0.01750000 0.00500000
#> V_VB V_RC V_R V_max R_oral R_IV
#> 0.01250000 0.82000000 0.20500000 3.53553391 0.01041667 0.00000000We can examine the initial conditions for the state variables in the model using the following command.
pbpk_mod$Y0
#> A_G A_L A_F A_AB A_VB A_R A_dose A_m AUC
#> 0 0 0 0 0 0 0 0 0As stated in the model specification file,
pbpk_simple.model, the default values for the initial
conditions are all zero.
Suppose that we want to perform a simulation and generate results for 100 hours following oral administration with an output resolution of 0.1 hours. We can create a vector of desired output times (i.e., ) using the following command.
times <- seq(from = 0, to = 100, by = 0.1)Then, we can perform the simulation and store the results in a
“matrix” data structure called out_oral as follows.
out_oral <- pbpk_mod$runModel(times)Finally, we can plot the total amount lost vs. time using the following command.
plot(out_oral[, "time"], out_oral[, "A_tot"],
type = "l", lty = 1, lwd = 2,
xlab = "Time (h)", ylab = "Amount (mg)"
)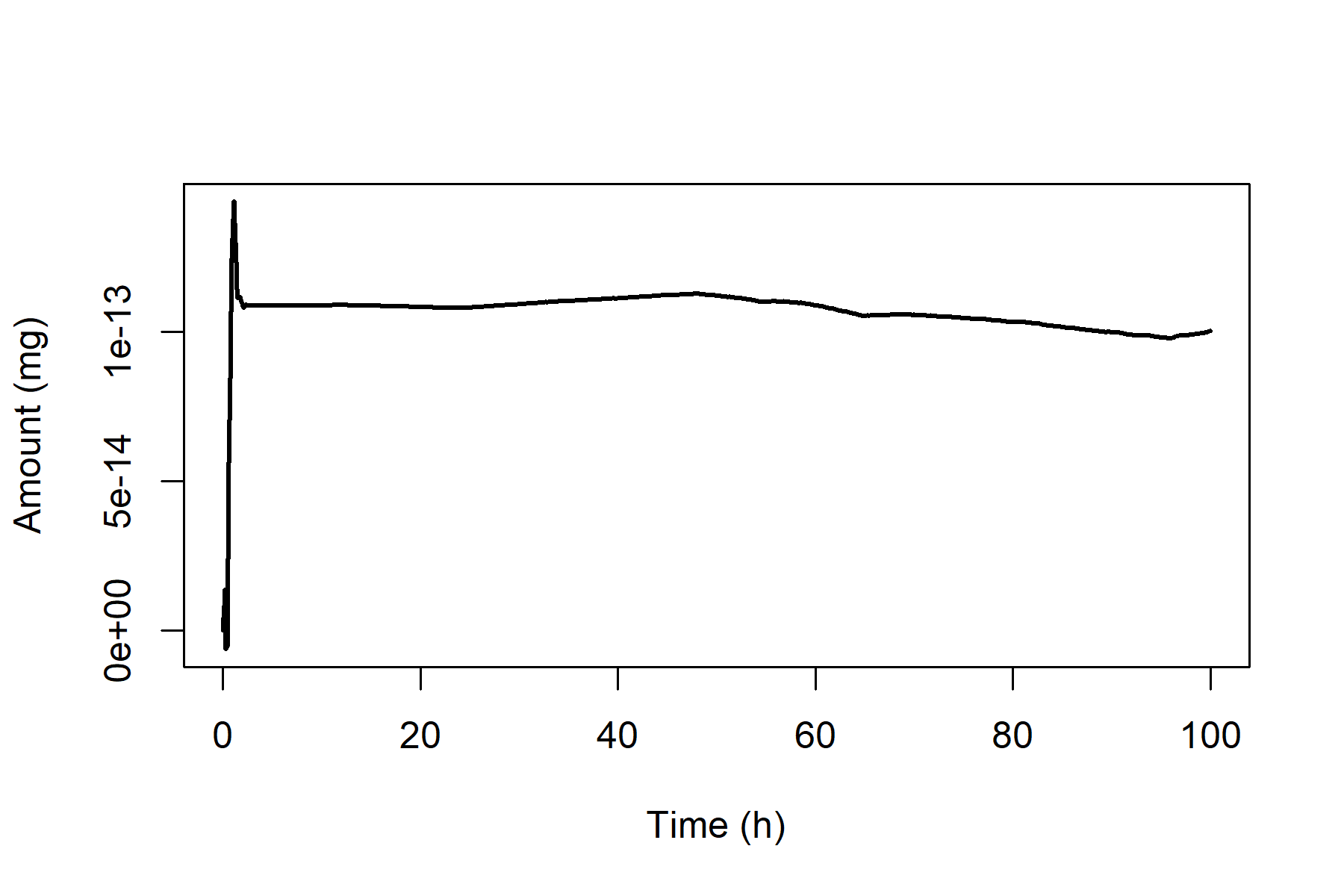 Note that the total amount lost is close to zero (less than
10)
at all time points examined. The degree of “closeness” to zero is
determined by the numerical precision of the ODE solver, which can be
adjusted using the arguments
Note that the total amount lost is close to zero (less than
10)
at all time points examined. The degree of “closeness” to zero is
determined by the numerical precision of the ODE solver, which can be
adjusted using the arguments rtol and atol
passed through runModel() to the deSolve
function ode. Further details about these arguments can be
found the documentation for the deSolve package.
Verifying Conservation of Flow
We can also verify that the total blood flow to the non-blood compartments in our PBPK model equals the total cardiac output using the following commands.
Examining the Simulation Results
After performing a simulation and storing the results (in this case,
in out_oral), we can create visual representations of these
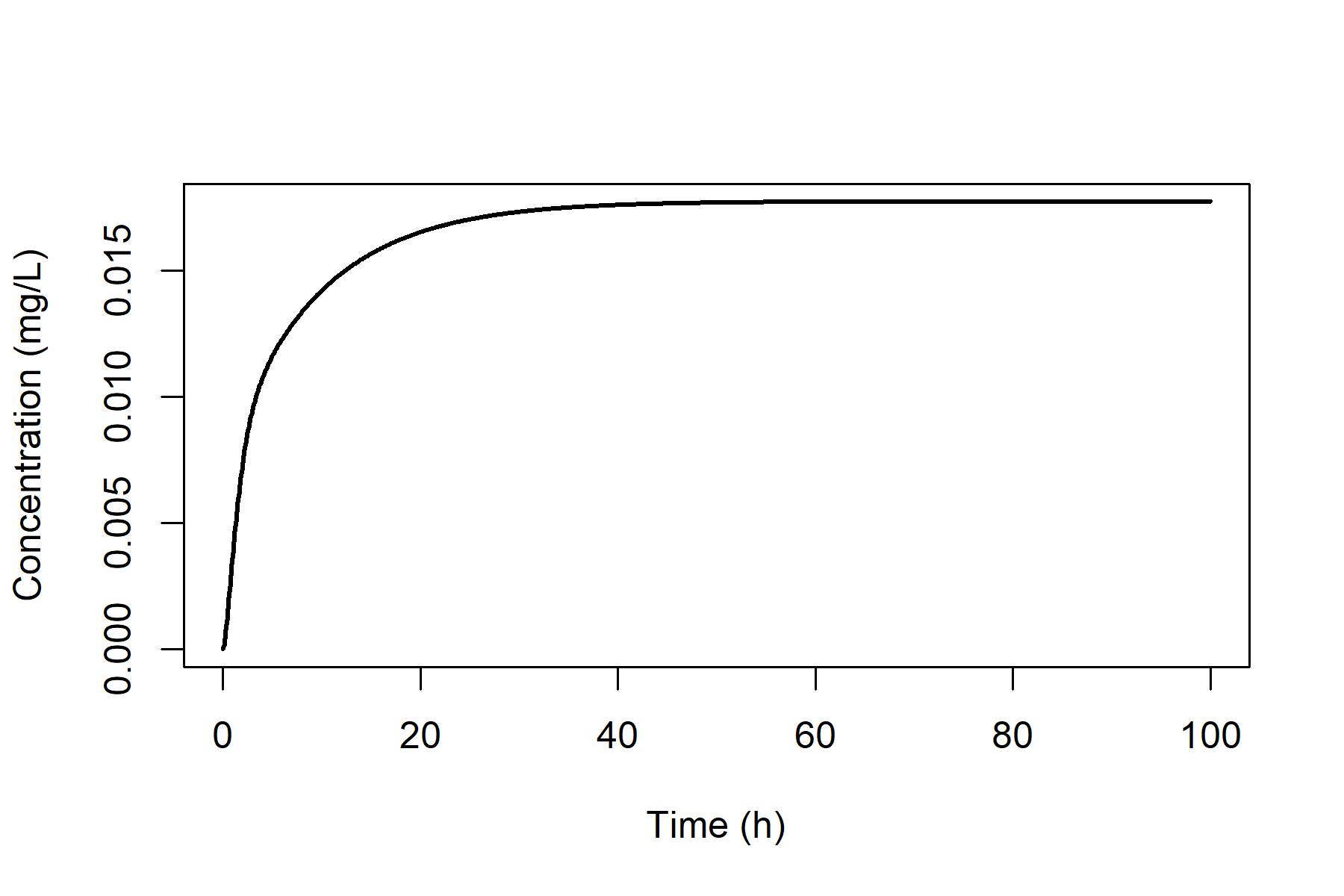
results. For example, we can plot the venous blood concentration
vs. time using the following commands.
plot(out_oral[, "time"], out_oral[, "C_VB"],
type = "l", lty = 1, lwd = 2,
xlab = "Time (h)", ylab = "Concentration (mg/L)"
)
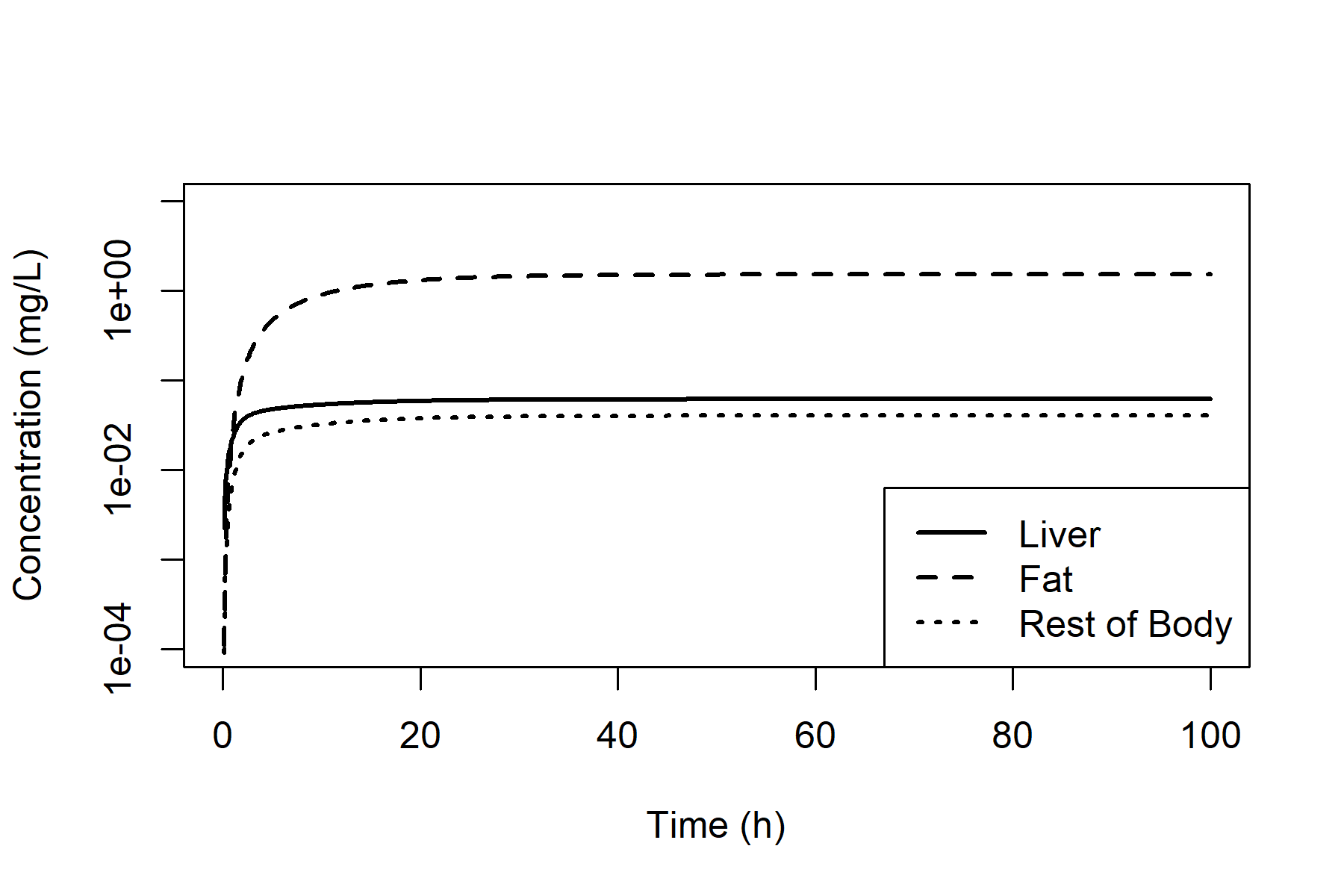
We can plot the concentrations in all non-blood compartments (on a logarithmic scale) as follows.
plot(out_oral[, "time"], out_oral[, "C_L"],
type = "l", lty = 1, lwd = 2,
log = "y", ylim = c(0.0001, 10),
xlab = "Time (h)", ylab = "Concentration (mg/L)"
)
lines(out_oral[, "time"], out_oral[, "C_F"],
lty = 2, lwd = 2
)
lines(out_oral[, "time"], out_oral[, "C_R"],
lty = 3, lwd = 2
)
legend("bottomright",
legend = c("Liver", "Fat", "Rest of Body"),
lty = c(1, 2, 3), lwd = 2
)