Cover Types
VELMA version 2.0 includes two major changes to the vegetation submodel:
- Multiple cover types (forest, grassland, agricultural crops, etc.) within a watershed can now be modeled. Theversion 1.0 convention of one cover type per grid cell is maintained.
- Plant biomass for each cover type is modeled using four plant tissues - leaves, aboveground stems,belowground stems, and roots - rather than as a single biomass pool.
Sections 5.1 - 5.3, below, discuss procedures for initializing a simulator configuration for one or more covertypes.
Sections 5.4 through 5.7, below, discuss procedures for initializing a simulator configuration for the four plantbiomass pools.
5.1 - Cover ID Map File
The VELMA program requires a set of Cover Species parameters for each Cover ID number that occurs in the Cover IDMap File. Each Cover ID corresponds to a distinct, user-defined Cover Species (cover type) - for example,hardwood forest, grassland, corn cropland, and so forth.
This section describes the Cover ID map file and associated parameter specifications.
The Cover ID Map File is specified by the coverSpeciesIndexMapFileName parameter. You can view and set itby selecting "All Parameters 5.1 Cover ID Map File" from the All Parameters outline drop-down menu:
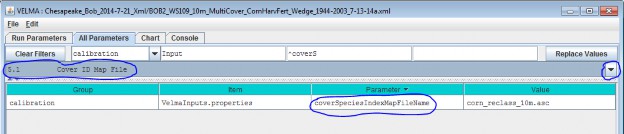
The file specified for the coverSpeciesIndexMapFileName is assumed to be a grid ASCII (".asc") map filewith the same row and column dimensions as the simulation configuration's DEM file. Its contents should beinteger values. Each cell's integer value should be the ID number of a cover type.
Suppose the specified cover index map file contains three distinct integers: 5, 7 and 9. For this map, the VELMAsimulation will expect the simulation configuration to include 3 Cover Species.
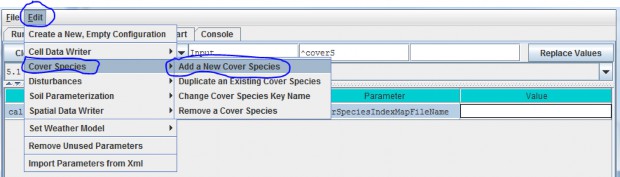
You will need to add a new Cover Species to a simulation configuration using the following steps:
- Click Edit Cover Species "Add a New Cover Species" menu item. This will cause a pop-up menuto appear (see step 2).
5.3 - Cover IDs and Names
[Note: All Parameters 5.2 - Cover Age Map File has been moved below section 5.3 to reflect therequired workflow]
Picking up from All Parameters 5.2, step (2)…
As an example: Suppose we are adding the first Cover Species ("5") for the hypothetical Cover ID Map File mentioned above, andthat ID "5" identifies cells that contain Conifers. Acceptable names (guaranteed unique because this is the first Cover Species we're adding) might be: "Conifer" or"Cover_5_Conifer", or something similar. However, the only acceptable ID value is "5" - because the Unique Cover ID # of this parameterization is whatkeys it to those particular cells in the Cover ID Map File.
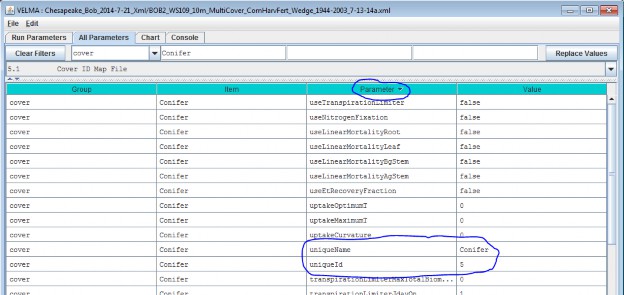
We did one other thing to get the above display: notice that the "Parameter" column is circled, and has a"down-arrow" indicating "descending-sort". After clicking OK to add the new Cover Species, we clicked the"Parameter" header field (twice), which sorts the table of properties on that column.
5.2 - Cover Age Map File
[Note: section 5.2 appears after section 5.3 to reflect the required workflow for developing a Cover Age Map fileand associated parameter values]
To enable scheduling of disturbance events during a simulation (per All Parameters section 24.0), VELMA needs tokeeps track of changes in the age (years) of vegetation from one calendar year to another, or as a result ofdisturbances that can reset stand age to 0 (e.g., by clearcutting) or to some other age (e.g., by selectivethinning of old trees that establishes understory vegetation as the new dominant age class). Note that cover agemaps can be can be applied to cover types other than forests, e.g., rangeland vegetation that may have a more diverse mixt ofspecies than vegetation recovering from a recent fire.
After you have established a Cover ID Map file (section 5.1) and specified Cover IDs and names for each CoverSpecies (section 5.3, above!), you will need to set up a Cover Age Map file and specify the file name.
The Cover Age Map file name is specified by the coverAgeMapFileName parameter. You can view and set it byselecting "All Parameters 5.2 Cover Age Map File" from the All Parameters outline drop-down menu:
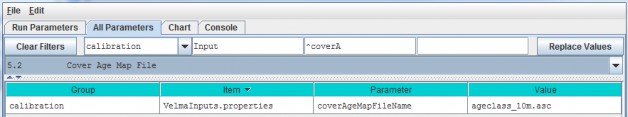
The file specified for the coverAgeMapFileName is assumed to be a grid ASCII (".asc") map file with thesame row and column dimensions as the simulation configuration's DEM file. Its contents should be integer valuesreflecting the age of vegetation (Cover Species) occupying each grid cell. Each cell's integer value should bethe ID number of a Cover Species. For example, an age of 0 could be assigned to row crops (corn, soybeans,etc.). Cells in a forest landscape might range in age from 0 (newly burned or planted) to many centuries. TheVELMA team has used biomass-to-age and tree height-to-age relationships to establish Cover Age Maps.
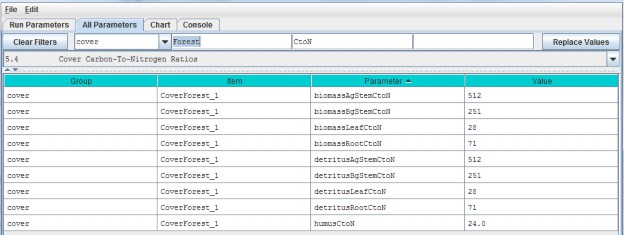
5.4 - Cover Carbon-To-Nitrogen Ratios
This section is concerned with initializing plant tissue C/N ratios for plant biomass and detritus associatedwith leaves, aboveground stems (AgStem), belowground stems (BgStem) and roots. Thus, a total of eight C/N (CtoN)ratios must be specified for each cover type. Only one cover type (CoverForest_1) is displayed for the examplebelow. In addition, a C/N ratio must be specified for the humus pool.
5.5 - Cover Uniform-Cell Initialization Amounts
Using the All Parameters drop-down menu, select "5.5 Cover Uniform-Cell Initialization Amounts". Theparameters shown are used to specify initial pool sizes (g C / m2) that VELMA
requires at the start of each simulation run. Values must be specified for all plant biomass pools, detrituspools and humus pool associated with each cover type. The example below shows just one cover type, but the samelist of 9 parameters repeats for each additional cover type that has been has been for the simulationconfiguration.
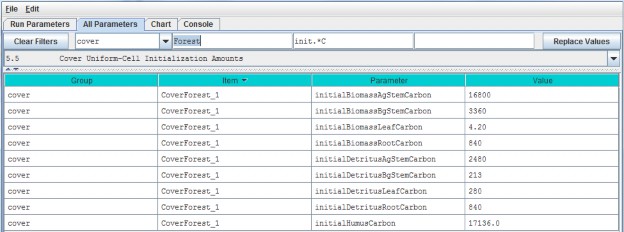
"Cover Uniform-Cell Initialization Amounts" refers to the fact that all grid cells assigned to a particular covertype will have the same initial values for plant biomass, detritus and humus. Obviously this method will notwork for watersheds where there is significant spatial variability in pool sizes (e.g., where forest stands varyin age). In these situations, you can use All Parameters menu item "24.3 Disturbance Items Spatial Specifiers"to establish spatially variable initialization amounts for any or all pools.
5.6 - Cover Gale-Grigal Root Parameter
Select section 5.6 from the All Parameters drop-down menu to specify Gale-Grigal root parameter values for eachcover type in your watershed:
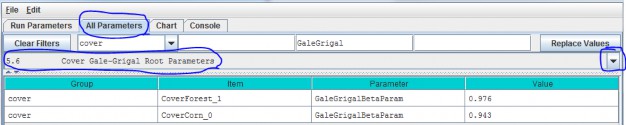
Parameter Definitions
| Parameter Name | Parameter Description |
|---|---|
| GaleGrigalBetaParam | The Beta term for this cover type's root vertical distribution. For the root distributionfunction of Gale Grigal (1987): roots[D] = 1 - B^D, where B = GaleGrigalBetaParam value (assigned here) D =layer depth |
References
Gale, M. R., & Grigal, D. F. (1987). Vertical root distributions of northern tree species in relation tosuccessional status. Canadian Journal of Forest Research, 17(8), 829-834.
Jackson, R. B., Canadell, J., Ehleringer, J. R., Mooney, H. A., Sala, O. E., & Schulze, E. D. (1996). Aglobal analysis of root distributions for terrestrial biomes. Oecologia, 108(3), 389-411.
Calibration Notes:
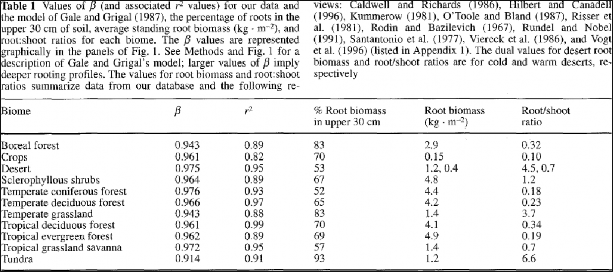
VELMA uses the method of Gale and Grigal (1987) to vertically distribute a cover type's specified total root biomass. This results in an exponentially decreasing amount of root biomass per soil layer. See Appendix 6.2 section A1.2.6 (Abdelnour et al. 2013) for a description of VELMA's implementation of the Gale-Grigal root distribution function.
Jackson et al. (1996) used the Gale-Grigal method to characterize rooting patterns for terrestrial biomesglobally. In the absence of measured data to calibrate GaleGrigalBetaParam for your site's cover types,we recommend consulting Jackson et al.'s Table 1and Figure 1:
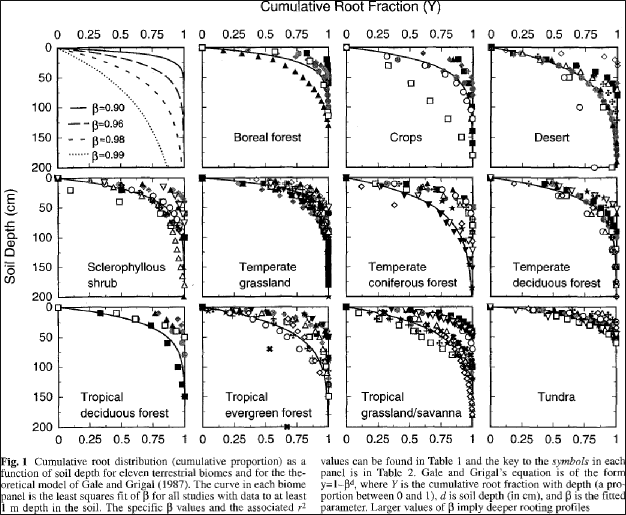
5.7 - Cover Leaf Stem and Root Pool Parameters
Select section 5.7 from the All Parameters drop-down menu to specify leaf stem and root pool parameter values foreach cover type in your watershed (do this for each cover type):
Parameters in this section cover the Leaf, Stem and Root pool C/N ratios, initial values, mortalityparameterizations and uptake fractions. Because each biomass or detritus leaf, stem and root pool has its ownset of parameters for these subjects, this list of parameters is relatively extensive.
Parameter Definitions
| Parameter Name | Parameter Description |
|---|---|
| biomassLeafCtoN | The Carbon-to-Nitrogen ratio of the cover type's Leaf biomass pool. |
| biomassAgStemCtoN | The Carbon-to-Nitrogen ratio of the cover type's AgStem biomass pool. |
| biomassBgStemCtoN | The Carbon-to-Nitrogen ratio of the cover type's BgStem biomass pool. |
| biomassRootCtoN | The Carbon-to-Nitrogen ratio of the cover type's Root biomass pool. (All layers ofthis pool use the same CtoN ratio value.) |
| detritusLeafCtoN | The Carbon-to-Nitrogen ratio of the cover type's Leaf detritus pool. |
| detritusAgStemCtoN | The Carbon-to-Nitrogen ratio of the cover type's AgStem detritus pool. |
| detritusBgStemCtoN | The Carbon-to-Nitrogen ratio of the cover type's BgStem detritus pool. (Alllayers of this pool use the same CtoN ratio value.) |
| detritusRootCtoN | The Carbon-to-Nitrogen ratio of the cover type's Root detritus pool. (All layersof this pool use the same CtoN ratio value.) |
| detritusLeafNmaxDecay | Maximum decay rate for Potter decomposition of Leaf detritus pool's Nitrogen amount. |
| detritusAgStemNmaxDecay | Maximum decay rate for Potter decomposition of AgStem detritus pool's Nitrogen amount. |
| detritusBgStemNmaxDecay | Maximum decay rate for Potter decomposition of BgStem detritus pool's Nitrogen amount. |
| detritusRootNmaxDecay | Maximum decay rate for Potter decomposition of Root detritus pool's Nitrogen amount. |
| initialBiomassLeafCarbon | The amount of biomass (in grams of Carbon)placed in the Leaf biomass pool of each cell of this cover type at initialization start. |
| initialBiomassAgStemCarbon | The amount of biomass (in grams of Carbon) placed in the AgStem biomass pool of each cell of this cover type at initialization start. |
| initialBiomassBgStemCarbon | The amount of biomass (in grams of Carbon)placed in the BgStem biomass pool of each cell of this cover type at initialization start. |
| initialBiomassRootCarbon | The amount of biomass (in grams of Carbon) placed in the Root biomass pool of each cell of this cover type at initialization start. |
| initialDetritusLeafCarbon | The amount of detritus (in grams of Carbon) placed in the Leaf detritus pool of each cell of this cover type at initialization start. |
| initialDetritusAgStemCarbon | The amount of detritus (in grams of Carbon) placed in the AgStem detritus pool of each cell of this cover type at initialization start. |
| initialDetritusBgStemCarbon | The amount of detritus (in grams of Carbon) placed in the BgStem detritus pool of each cell of this cover type at initialization start. |
| initialDetritusRootCarbon | The amount of detritus (in grams of Carbon) placed in the Root detritus pool of each cell of this cover type at initialization start. |
| etRecoveryFractionMinimumLeafBiomassC | The minimum amount of leaf biomass (in gC/m^2) required to compute an ET recovery fraction. Whenever leaf biomass is less than this amount, the ET recoveryfraction is forced to the etRecoverFractionMinimumValue. See Section 10.2.1 ET Recovery Parameters. |
| mortalityLeafAnnualFraction | The fraction of the Leaf biomass Pool annual mortality amount actually subtracted from that pool per day of senescence. The default value for this fraction is1.0 (i.e. the full per-day amount) and the valid range is [0.0 to 1.0]. Warning! Users unfamiliar with thisparameter's behavior should leave it set to the default value of 1.0. |
| mortalityLeafDenominatorCoefficient | The mortality equation's denominator coefficient value for the biomass Leaf pool. |
| mortalityLeafExponentialCoefficient | The mortality equation's exponential coefficient value for the biomass Leaf pool. |
| mortalityLeafNumeratorCoefficient | The mortality equation's numerator coefficient value for biomass Leaf pool. |
| mortalityAgStemAnnualFraction | The fraction of the AgStem biomass Pool annual mortality amount actually subtracted from that pool per day of senescence. The default value for this fraction is1.0 (i.e. the full per-day amount) and the valid range is [0.0 to 1.0]. Warning! Users unfamiliar with thisparameter's behavior should leave it set to the default value of 1.0. |
| mortalityAgStemDenominatorCoefficient | The mortality equation's denominator coefficient value for the biomass AgStem pool. |
| mortalityAgStemExponentialCoefficient | The mortality equation's exponential coefficient value for the biomass AgStem pool. |
| mortalityAgStemNumeratorCoefficient | The mortality equation's numerator coefficient value for the biomass AgStem pool. |
| mortalityBgStemAnnualFraction | The fraction of the BgStem biomass Pool annual mortality amount actually subtracted from that pool per day of senescence. The default value for this fraction is1.0 (i.e. the full per-day amount) and the valid range is [0.0 to 1.0]. Warning! Users unfamiliar with thisparameter's behavior should leave it set to the default value of 1.0. |
| mortalityBgStemDenominatorCoefficient | The mortality equation's denominator coefficient value for the biomass BgStem pool. |
| mortalityBgStemExponentialCoefficient | The mortality equation's exponential coefficient value for the biomass BgStem pool. |
| mortalityBgStemNumeratorCoefficient | The mortality equation's numerator coefficient value for the biomass BgStem pool. |
| mortalityRootAnnualFraction | The fraction of the Leaf biomass Pool annual mortality amount actually subtracted from that pool per day of senescence. The default value for this fraction is1.0 (i.e. the full per-day amount) and the valid range is [0.0 to 1.0]. Warning! Users unfamiliar with thisparameter's behavior should leave it set to the default value of 1.0. |
| mortalityRootDenominatorCoefficient | The mortality equation's denominator coefficient value for the biomass Root pool. |
| mortalityRootExponentialCoefficient | The mortality equation's exponential coefficient value for the biomass Root pool. |
| mortalityRootNumeratorCoefficient | The mortality equation's numerator coefficient value for the biomass Root pool. |
| useLinearMortalityLeaf | When true, leaf mortality is computed using a simple linear function. When false (default)mortality is computed via a logistic function. |
| useLinearMortalityAgStem | When true, AgStem mortality is computed using a simple linear function. When false (default) mortality is computed via a logistic function. |
| useLinearMortalityBgStem | When true, BgStem mortality is computed using a simple linear function. When false (default) mortality is computed via a logistic function. |
| useLinearMortalityRoot | When true, Root mortality is computed using a simple linear function. When false (default)mortality is computed via a logistic function. |
| nppToBiomassLeafNfraction | The fraction of total daily NPP for a cell of this cover type allotted to the biomass Leaf pool. |
| nppToBiomassRootNfraction | The fraction of total daily NPP for a cell of this cover type allotted to the biomass Root pool. |
| nppToBiomassAgStemNfraction | The fraction of daily NPP ALOTTED TO STEM POOLS for a cell of this cover type allotted to the biomass AgStem pool. The remaining NPP allotted to stem pools goes to the biomass BgStem pool. NOTE: Total dailyNPP is reduced by the fractions for leaf and root pools first. Any remainder is the daily NPP allotted to stempools. |
Calibration Notes
At this stage (section 5.7) of your simulation configuration, don't try to specify values for all the parameterslisted above. For now, just focus on the initial C and N pools for plant biomass anddetritus (highlighted in yellow, above). To specify initial C and N stocks(g/m2) for these pools, we recommend that you consultAppendix 1 for an overview of VELMA's LSR submodel and it C and N pools and fluxes.Filling in C and N values for the various pools shown would be a good start. Remember to document data sourcesas you go.
Methods for specifying parameter values for NPP (plant uptake) allocation, plant biomass mortality, and detritusdecay (decomposition) rates, and NPP are discussed in subsequent sections:
- N uptake (NPP): Section 11.0
- Mortality: Section 12.0
- Decomposition: Section 13.0
5.7.1 - Cover Leaf Pool Parameter
Select section 5.7.1 from the All Parameters drop-down menu to specify cover-specific parameter values for leafbiomass and detritus. See 5.7 above for the parameter descriptions.
5.7.2 - Cover Above-Ground Stem Pool Parameters
Select section 5.7.1 from the All Parameters drop-down menu to specify cover-specific parameter values foraboveground stem biomass and detritus. See 5.7 above for the parameter descriptions.
5.7.3 - Cover Below-Ground Stem Pool Parameters
Select section 5.7.1 from the All Parameters drop-down menu to specify cover-specific parameter values forbelowground stem biomass and detritus. See 5.7 above for the parameter descriptions.
5.7.4 - Cover Root Pool Parameters
Select section 5.7.1 from the All Parameters drop-down menu to specify cover-specific parameter values for rootbiomass and detritus. See 5.7 above for the parameter descriptions.