Cape Cod - Detailed step-by-step
Standardize, clean and wrangle Water Quality Portal data in Cape Cod into more analytic-ready formats using the harmonize_wq package
US EPA’s Water Quality Portal (WQP) aggregates water quality, biological, and physical data provided by many organizations and has become an essential resource with tools to query and retrieval data using python or R. Given the variety of data and variety of data originators, using the data in analysis often requires data cleaning to ensure it meets the required quality standards and data wrangling to get it in a more analytic-ready format. Recognizing the definition of analysis-ready varies depending on the analysis, the harmonixe_wq package is intended to be a flexible water quality specific framework to help:
Identify differences in data units (including speciation and basis)
Identify differences in sampling or analytic methods
Resolve data errors using transparent assumptions
Reduce data to the columns that are most commonly needed
Transform data from long to wide format
Domain experts must decide what data meets their quality standards for data comparability and any thresholds for acceptance or rejection.
Detailed step-by-step workflow
This example workflow takes a deeper dive into some of the expanded functionality to examine results for different water quality parameters in Cape Cod
Install and import the required libraries
[1]:
import sys
#! python -m pip uninstall harmonize-wq --yes
# Use pip to install the package from pypi or the latest from github
#!{sys.executable} -m pip install harmonize-wq
# For latest dev version
#!{sys.executable} -m pip install git+https://github.com/USEPA/harmonize-wq.git
[2]:
import dataretrieval.wqp as wqp
from harmonize_wq import wrangle
from harmonize_wq import location
from harmonize_wq import harmonize
from harmonize_wq import visualize
from harmonize_wq import clean
Download location data using dataretrieval
[3]:
# Read geometry for Area of Interest from geojson file url and plot
aoi_url = 'https://github.com/jbousquin/test_notebook/raw/main/temperature_data/NewEngland.geojson'
aoi_gdf = wrangle.as_gdf(aoi_url) # Already 4326 standard
aoi_gdf.plot()
[3]:
<Axes: >
[4]:
# Build query with characteristicNames and the AOI extent
query = {'characteristicName': ['Phosphorus',
'Temperature, water',
'Depth, Secchi disk depth',
'Dissolved oxygen (DO)',
'Salinity',
'pH',
'Nitrogen',
'Conductivity',
'Organic carbon',
'Chlorophyll a',
'Turbidity',
'Sediment',
'Fecal Coliform',
'Escherichia coli']}
query['bBox'] = wrangle.get_bounding_box(aoi_gdf)
[5]:
# Query stations (can be slow)
stations, site_md = wqp.what_sites(**query)
[6]:
# Rows and columns for results
stations.shape
[6]:
(11312, 37)
[7]:
# First 5 rows
stations.head()
[7]:
| OrganizationIdentifier | OrganizationFormalName | MonitoringLocationIdentifier | MonitoringLocationName | MonitoringLocationTypeName | MonitoringLocationDescriptionText | HUCEightDigitCode | DrainageAreaMeasure/MeasureValue | DrainageAreaMeasure/MeasureUnitCode | ContributingDrainageAreaMeasure/MeasureValue | ... | AquiferName | LocalAqfrName | FormationTypeText | AquiferTypeName | ConstructionDateText | WellDepthMeasure/MeasureValue | WellDepthMeasure/MeasureUnitCode | WellHoleDepthMeasure/MeasureValue | WellHoleDepthMeasure/MeasureUnitCode | ProviderName | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | USGS-MA | USGS Massachusetts Water Science Center | USGS-010965305 | MERRIMACK R NR TYNGSBOROUGH BRIDGE TYNGSBOROUG... | Stream | NaN | 1070006.0 | 4070.00 | sq mi | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NWIS |
| 1 | USGS-MA | USGS Massachusetts Water Science Center | USGS-01096544 | STONY BROOK AT SCHOOL STREET AT CHELMSFORD, MA | Stream | NaN | 1070006.0 | 41.57 | sq mi | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NWIS |
| 2 | USGS-MA | USGS Massachusetts Water Science Center | USGS-01096546 | STONY BROOK AT CHELMSFORD, MA | Stream | NaN | 1070006.0 | 43.60 | sq mi | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NWIS |
| 3 | USGS-MA | USGS Massachusetts Water Science Center | USGS-01096548 | STONY BROOK AT N CHELMSFORD, MA | Stream | NaN | 1070006.0 | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NWIS |
| 4 | USGS-MA | USGS Massachusetts Water Science Center | USGS-01096550 | MERRIMACK RIVER ABOVE LOWELL, MA | Stream | NaN | 1070006.0 | 3900.00 | sq mi | 3900.0 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NWIS |
5 rows × 37 columns
[8]:
# Columns used for an example row
stations.iloc[0][['HorizontalCoordinateReferenceSystemDatumName', 'LatitudeMeasure', 'LongitudeMeasure']]
[8]:
HorizontalCoordinateReferenceSystemDatumName NAD83
LatitudeMeasure 42.677389
LongitudeMeasure -71.421056
Name: 0, dtype: object
[9]:
# Harmonize location datums to 4326 (Note we keep intermediate columns using intermediate_columns=True)
stations_gdf = location.harmonize_locations(stations, outEPSG=4326, intermediate_columns=True)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/clean.py:356: FutureWarning: Logical ops (and, or, xor) between Pandas objects and dtype-less sequences (e.g. list, tuple) are deprecated and will raise in a future version. Wrap the object in a Series, Index, or np.array before operating instead.
cond_notna = mask & (df_out["QA_flag"].notna()) # Mask cond and not NA
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/clean.py:360: FutureWarning: Logical ops (and, or, xor) between Pandas objects and dtype-less sequences (e.g. list, tuple) are deprecated and will raise in a future version. Wrap the object in a Series, Index, or np.array before operating instead.
df_out.loc[mask & (df_out["QA_flag"].isna()), "QA_flag"] = flag
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/clean.py:360: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value 'LatitudeMeasure: Imprecise: lessthan3decimaldigits' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[mask & (df_out["QA_flag"].isna()), "QA_flag"] = flag
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/clean.py:356: FutureWarning: Logical ops (and, or, xor) between Pandas objects and dtype-less sequences (e.g. list, tuple) are deprecated and will raise in a future version. Wrap the object in a Series, Index, or np.array before operating instead.
cond_notna = mask & (df_out["QA_flag"].notna()) # Mask cond and not NA
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/clean.py:360: FutureWarning: Logical ops (and, or, xor) between Pandas objects and dtype-less sequences (e.g. list, tuple) are deprecated and will raise in a future version. Wrap the object in a Series, Index, or np.array before operating instead.
df_out.loc[mask & (df_out["QA_flag"].isna()), "QA_flag"] = flag
[10]:
# Every function has a dostring to help understand input/output and what it does
location.harmonize_locations?
[11]:
# Rows and columns for results after running the function (5 new columns, only 2 new if intermediate_columns=False)
stations_gdf.shape
[11]:
(11312, 42)
[12]:
# Example results for the new columns
stations_gdf.iloc[0][['geom_orig', 'EPSG', 'QA_flag', 'geom', 'geometry']]
[12]:
geom_orig (-71.4210556, 42.67738889)
EPSG 4269.0
QA_flag NaN
geom POINT (-71.4210556 42.67738889)
geometry POINT (-71.4210556 42.67738889)
Name: 0, dtype: object
[13]:
# geom and geometry look the same but geometry is a special datatype
stations_gdf['geometry'].dtype
[13]:
<geopandas.array.GeometryDtype at 0x7faff6088310>
[14]:
# Look at the different QA_flag flags that have been assigned,
# e.g., for bad datums or limited decimal precision
set(stations_gdf.loc[stations_gdf['QA_flag'].notna()]['QA_flag'])
[14]:
{'HorizontalCoordinateReferenceSystemDatumName: Bad datum OTHER, EPSG:4326 assumed',
'HorizontalCoordinateReferenceSystemDatumName: Bad datum UNKWN, EPSG:4326 assumed',
'LatitudeMeasure: Imprecise: lessthan3decimaldigits',
'LatitudeMeasure: Imprecise: lessthan3decimaldigits; LongitudeMeasure: Imprecise: lessthan3decimaldigits',
'LongitudeMeasure: Imprecise: lessthan3decimaldigits'}
[15]:
# Map it
stations_gdf.plot()
[15]:
<Axes: >
[16]:
# Clip to area of interest
stations_clipped = wrangle.clip_stations(stations_gdf, aoi_gdf)
[17]:
# Map it
stations_clipped.plot()
[17]:
<Axes: >
[18]:
# How many stations now?
len(stations_clipped)
[18]:
1880
[19]:
# To save the results to a shapefile
#import os
#path = '' #specify the path (folder/directory) to save it to
#stations_clipped.to_file(os.path.join(path,'CapeCod_stations.shp'))
Retrieve Characteristic Data
[20]:
# Now query for results
query['dataProfile'] = 'narrowResult'
res_narrow, md_narrow = wqp.get_results(**query)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/dataretrieval/wqp.py:153: DtypeWarning: Columns (8,10,13,15,17,19,20,21,22,23,28,31,33,34,36,38,60,63,64,65,66,67,68,69,70,71,72) have mixed types. Specify dtype option on import or set low_memory=False.
df = pd.read_csv(StringIO(response.text), delimiter=",")
[21]:
df = res_narrow
df
[21]:
| OrganizationIdentifier | OrganizationFormalName | ActivityIdentifier | ActivityStartDate | ActivityStartTime/Time | ActivityStartTime/TimeZoneCode | MonitoringLocationIdentifier | ResultIdentifier | DataLoggerLine | ResultDetectionConditionText | ... | AnalysisEndTime/TimeZoneCode | ResultLaboratoryCommentCode | ResultLaboratoryCommentText | ResultDetectionQuantitationLimitUrl | LaboratoryAccreditationIndicator | LaboratoryAccreditationAuthorityName | TaxonomistAccreditationIndicator | TaxonomistAccreditationAuthorityName | LabSamplePreparationUrl | ProviderName | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | BRC | Blackstone River Coalition (Volunteer) | BRC-C-02-02-020:20131012040800:FM:0.2083333333... | 2013-10-12 | 04:08:00 | EST | BRC-C-02-02-020 | STORET-1039097035 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | STORET |
| 1 | OARS | OARS - For the Assabet Sudbury & Concord River... | OARS-CND-161:20130818:0651:FM:1 | 2013-08-18 | 06:51:00 | EDT | OARS-CND-161 | STORET-838568413 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | STORET |
| 2 | OARS | OARS - For the Assabet Sudbury & Concord River... | OARS-SUD-064:20130721:0610:FM:1 | 2013-07-21 | 06:10:00 | EDT | OARS-SUD-064 | STORET-838568309 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | STORET |
| 3 | CRWA | Charles River Watershed Association (Massachus... | CRWA-CYN20130809ROBTemp01 | 2013-08-09 | 11:14:33 | EST | CRWA-ROB | STORET-591631481 | 130809111433.0 | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | STORET |
| 4 | WTGHA | Wompanoag Tribe of Gay Head Aquinnah (Tribal) | WTGHA-m41:20131017:FM:B | 2013-10-17 | NaN | NaN | WTGHA-M41 | STORET-1041401688 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | STORET |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 574706 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_DUCK_W_19560816_731708 | 1956-08-16 | NaN | NaN | 11NPSWRD_WQX-CACO_DUCK_W | STORET-740598866 | NaN | NaN | ... | NaN | NaN | NaN | https://www.waterqualitydata.us/data/providers... | NaN | NaN | NaN | NaN | NaN | STORET |
| 574707 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_DUCK_W_19560616_731704 | 1956-06-16 | NaN | NaN | 11NPSWRD_WQX-CACO_DUCK_W | STORET-740598862 | NaN | NaN | ... | NaN | NaN | NaN | https://www.waterqualitydata.us/data/providers... | NaN | NaN | NaN | NaN | NaN | STORET |
| 574708 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_DUCK_W_19560701_731705 | 1956-07-01 | NaN | NaN | 11NPSWRD_WQX-CACO_DUCK_W | STORET-740598863 | NaN | NaN | ... | NaN | NaN | NaN | https://www.waterqualitydata.us/data/providers... | NaN | NaN | NaN | NaN | NaN | STORET |
| 574709 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_GREAT_W_19550816_731703 | 1955-08-16 | NaN | NaN | 11NPSWRD_WQX-CACO_GREAT_W | STORET-740649462 | NaN | NaN | ... | NaN | NaN | NaN | https://www.waterqualitydata.us/data/providers... | NaN | NaN | NaN | NaN | NaN | STORET |
| 574710 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_SLOUGH_19520816_731702 | 1952-08-16 | NaN | NaN | 11NPSWRD_WQX-CACO_SLOUGH | STORET-740745800 | NaN | Not Detected | ... | NaN | NaN | NaN | https://www.waterqualitydata.us/data/providers... | NaN | NaN | NaN | NaN | NaN | STORET |
574711 rows × 78 columns
[22]:
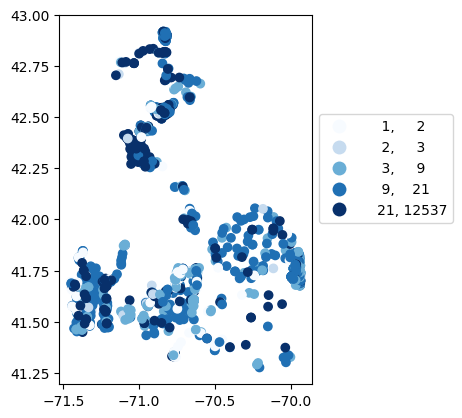
# Map number of usable results at each station
gdf_count = visualize.map_counts(df, stations_clipped)
legend_kwds = {"fmt": "{:.0f}", 'bbox_to_anchor':(1, 0.75)}
gdf_count.plot(column='cnt', cmap='Blues', legend=True, scheme='quantiles', legend_kwds=legend_kwds)
[22]:
<Axes: >
Harmonize Characteristic Results
Two options for functions to harmonize characteristics: harmonize_all() or harmonize_generic(). harmonize_all runs functions on all characteristics and lets you specify how to handle errors harmonize_generic runs functions only on the characteristic specified with char_val and lets you also choose output units, to keep intermediate columns and to do a quick report summarizing changes.
[23]:
# See Documentation
#harmonize_WQP.harmonize_all?
#harmonize_WQP.harmonize?
secchi disk depth
[24]:
# Each harmonize function has optional params, e.g., char_val is the characticName column value to use so we can send the entire df.
# Optional params: units='m', char_val='Depth, Secchi disk depth', out_col='Secchi', report=False)
# We start by demonstrating on secchi disk depth (units default to m, keep intermediate fields, see report)
df = harmonize.harmonize(df, "Depth, Secchi disk depth", errors="ignore", intermediate_columns=True, report=True)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/clean.py:360: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value 'ResultMeasureValue: missing (NaN) result' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[mask & (df_out["QA_flag"].isna()), "QA_flag"] = flag
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: '%' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(3.25, 'meter')> <Quantity(4.25, 'meter')>
<Quantity(3.5, 'meter')> ... <Quantity(17.0, 'meter')>
<Quantity(16.0, 'meter')> <Quantity(7.8, 'meter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
-Usable results-
count 8961.000000
mean -5.920774
std 298.744632
min -9999.000000
25% 1.400000
50% 2.000000
75% 3.962400
max 27.000000
dtype: float64
Unusable results: 2231
Usable results with inferred units: 8
Results outside threshold (0.0 to 1786.54702135132): 16
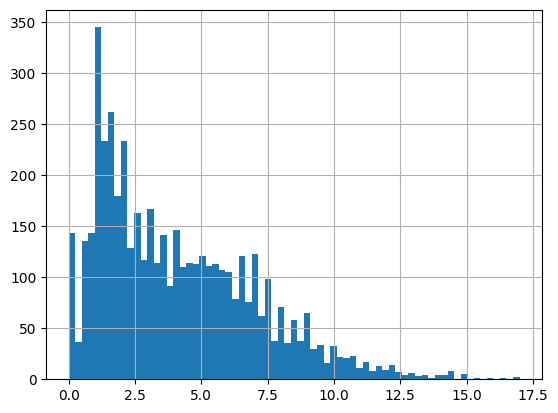
The threshold is based on standard deviations and is currently only used in the histogram.
[25]:
# Look at a table of just Secchi results and focus on subset of columns
cols = ['MonitoringLocationIdentifier', 'ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Units']
sechi_results = df.loc[df['CharacteristicName']=='Depth, Secchi disk depth', cols + ['Secchi']]
sechi_results
[25]:
| MonitoringLocationIdentifier | ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Units | Secchi | |
|---|---|---|---|---|---|---|
| 49 | 11113300-GRTKINSD | 3.25 | m | NaN | m | 3.25 meter |
| 179 | 11113300-BEADERD | 4.25 | m | NaN | m | 4.25 meter |
| 317 | 11113300-ANGSDND | 3.5 | m | NaN | m | 3.5 meter |
| 698 | 11113300-GRTKINSD | 2.25 | m | NaN | m | 2.25 meter |
| 741 | 11113300-GRTKINSD | 4.75 | m | NaN | m | 4.75 meter |
| ... | ... | ... | ... | ... | ... | ... |
| 574706 | 11NPSWRD_WQX-CACO_DUCK_W | 10.0 | m | NaN | m | 10.0 meter |
| 574707 | 11NPSWRD_WQX-CACO_DUCK_W | 17.0 | m | NaN | m | 17.0 meter |
| 574708 | 11NPSWRD_WQX-CACO_DUCK_W | 16.0 | m | NaN | m | 16.0 meter |
| 574709 | 11NPSWRD_WQX-CACO_GREAT_W | 7.8 | m | NaN | m | 7.8 meter |
| 574710 | 11NPSWRD_WQX-CACO_SLOUGH | NaN | m | ResultMeasureValue: missing (NaN) result | m | NaN |
11192 rows × 6 columns
[26]:
# Look at unusable(NAN) results
sechi_results.loc[df['Secchi'].isna()]
[26]:
| MonitoringLocationIdentifier | ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Units | Secchi | |
|---|---|---|---|---|---|---|
| 24555 | NARS_WQX-NLA12_MA-102 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | m | NaN |
| 38414 | 11113300-ISLDERD | =4.25 | m | ResultMeasureValue: "=4.25" result cannot be used | m | NaN |
| 38813 | 11113300-ROBHUDD | =2.25 | m | ResultMeasureValue: "=2.25" result cannot be used | m | NaN |
| 39063 | 11113300-ROBHUDD | =3 | m | ResultMeasureValue: "=3" result cannot be used | m | NaN |
| 39484 | 11113300-LONDVLD | =2.675 | m | ResultMeasureValue: "=2.675" result cannot be ... | m | NaN |
| ... | ... | ... | ... | ... | ... | ... |
| 495406 | 11113300-COBWINSD | =2.55 | m | ResultMeasureValue: "=2.55" result cannot be used | m | NaN |
| 495427 | 11113300-MILSALD | =2 | m | ResultMeasureValue: "=2" result cannot be used | m | NaN |
| 495441 | 11113300-LONPELNHD | =2.35 | m | ResultMeasureValue: "=2.35" result cannot be used | m | NaN |
| 498332 | 11113300-WORSALD | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | m | NaN |
| 574710 | 11NPSWRD_WQX-CACO_SLOUGH | NaN | m | ResultMeasureValue: missing (NaN) result | m | NaN |
2231 rows × 6 columns
[27]:
# look at the QA flag for first row from above
list(sechi_results.loc[df['Secchi'].isna()]['QA_flag'])[0]
[27]:
'ResultMeasureValue: missing (NaN) result; ResultMeasure/MeasureUnitCode: MISSING UNITS, m assumed'
[28]:
# All cases where there was a QA flag
sechi_results.loc[df['QA_flag'].notna()]
[28]:
| MonitoringLocationIdentifier | ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Units | Secchi | |
|---|---|---|---|---|---|---|
| 24555 | NARS_WQX-NLA12_MA-102 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | m | NaN |
| 38414 | 11113300-ISLDERD | =4.25 | m | ResultMeasureValue: "=4.25" result cannot be used | m | NaN |
| 38813 | 11113300-ROBHUDD | =2.25 | m | ResultMeasureValue: "=2.25" result cannot be used | m | NaN |
| 39063 | 11113300-ROBHUDD | =3 | m | ResultMeasureValue: "=3" result cannot be used | m | NaN |
| 39484 | 11113300-LONDVLD | =2.675 | m | ResultMeasureValue: "=2.675" result cannot be ... | m | NaN |
| ... | ... | ... | ... | ... | ... | ... |
| 495406 | 11113300-COBWINSD | =2.55 | m | ResultMeasureValue: "=2.55" result cannot be used | m | NaN |
| 495427 | 11113300-MILSALD | =2 | m | ResultMeasureValue: "=2" result cannot be used | m | NaN |
| 495441 | 11113300-LONPELNHD | =2.35 | m | ResultMeasureValue: "=2.35" result cannot be used | m | NaN |
| 498332 | 11113300-WORSALD | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | m | NaN |
| 574710 | 11NPSWRD_WQX-CACO_SLOUGH | NaN | m | ResultMeasureValue: missing (NaN) result | m | NaN |
1273 rows × 6 columns
If both value and unit are missing nothing can be done, a unitless (NaN) value is assumed as to be in default units but a QA_flag is added
[29]:
# Aggregate secchi data by station
visualize.station_summary(sechi_results, 'Secchi')
[29]:
| MonitoringLocationIdentifier | cnt | mean | |
|---|---|---|---|
| 0 | 11113300-ANGSDND | 45 | 3.765500 |
| 1 | 11113300-ARLSALD | 3 | 3.400000 |
| 2 | 11113300-BARKIND | 1 | 2.100000 |
| 3 | 11113300-BAYKIND | 1 | 1.900000 |
| 4 | 11113300-BEADERD | 46 | 3.666304 |
| ... | ... | ... | ... |
| 987 | WWMD_VA-SH1 | 2 | 1.500000 |
| 988 | WWMD_VA-SH2 | 2 | 1.800000 |
| 989 | WWMD_VA-SR6A | 1 | 0.500000 |
| 990 | WWMD_VA-WF2 | 1 | 0.600000 |
| 991 | WWMD_VA-WR5 | 1 | 2.000000 |
992 rows × 3 columns
[30]:
# Map number of usable results at each station
gdf_count = visualize.map_counts(sechi_results, stations_clipped)
gdf_count.plot(column='cnt', cmap='Blues', legend=True, scheme='quantiles', legend_kwds=legend_kwds)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/mapclassify/classifiers.py:1653: UserWarning: Not enough unique values in array to form 5 classes. Setting k to 4.
self.bins = quantile(y, k=k)
[30]:
<Axes: >
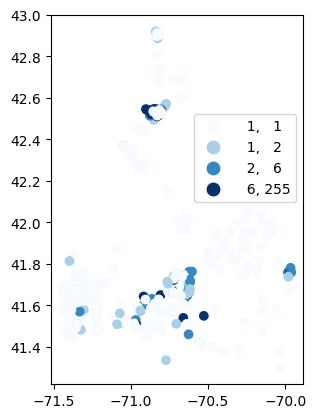
[31]:
# Map average results at each station
gdf_avg = visualize.map_measure(sechi_results, stations_clipped, 'Secchi')
gdf_avg.plot(column='mean', cmap='OrRd', legend=True)
[31]:
<Axes: >
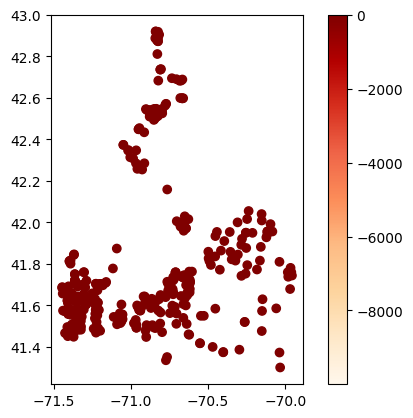
Temperature
The default error=’raise’, makes it so that there is an error when there is a dimensionality error (i.e. when units can’t be converted). Here we would get the error: DimensionalityError: Cannot convert from ‘count’ (dimensionless) to ‘degree_Celsius’ ([temperature])
[32]:
#'Temperature, water'
# errors=‘ignore’, invalid dimension conversions will return the NaN.
df = harmonize.harmonize(df, 'Temperature, water', intermediate_columns=True, report=True, errors='ignore')
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'count' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(22.18, 'degree_Celsius')> <Quantity(23.01, 'degree_Celsius')>
<Quantity(17.39, 'degree_Celsius')> ... <Quantity(4.0, 'degree_Celsius')>
<Quantity(1.5, 'degree_Celsius')> <Quantity(3.9, 'degree_Celsius')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
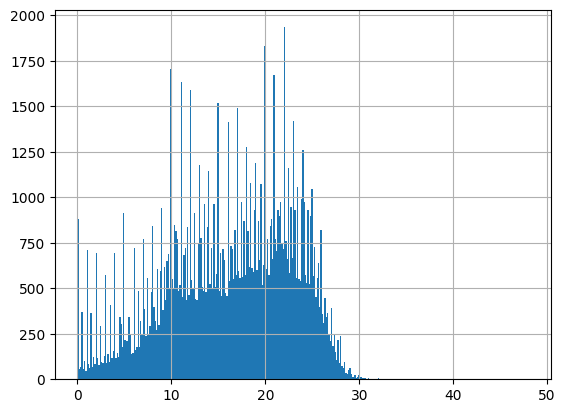
-Usable results-
count 150547.000000
mean 16.296038
std 7.274667
min -9.830000
25% 11.200000
50% 17.300000
75% 21.900000
max 910.000000
dtype: float64
Unusable results: 226
Usable results with inferred units: 217
Results outside threshold (0.0 to 59.94403905878173): 477
[33]:
# Look at what was changed
cols = ['MonitoringLocationIdentifier', 'ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Temperature', 'Units']
temperature_results = df.loc[df['CharacteristicName']=='Temperature, water', cols]
temperature_results
[33]:
| MonitoringLocationIdentifier | ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Temperature | Units | |
|---|---|---|---|---|---|---|
| 1 | OARS-CND-161 | 22.18 | deg C | NaN | 22.18 degree_Celsius | degC |
| 3 | CRWA-ROB | 23.01 | deg C | NaN | 23.01 degree_Celsius | degC |
| 5 | WTGHA-M43 | 17.39 | deg C | NaN | 17.39 degree_Celsius | degC |
| 8 | MASSDEP-W2386 | 20.9 | deg C | NaN | 20.9 degree_Celsius | degC |
| 12 | WTGHA-S35 | 22.54 | deg C | NaN | 22.54 degree_Celsius | degC |
| ... | ... | ... | ... | ... | ... | ... |
| 574691 | 11NPSWRD_WQX-CACO_GULL | 4.8 | deg C | NaN | 4.8 degree_Celsius | degC |
| 574692 | 11NPSWRD_WQX-CACO_GULL | 9.1 | deg C | NaN | 9.1 degree_Celsius | degC |
| 574693 | 11NPSWRD_WQX-CACO_GULL | 4.0 | deg C | NaN | 4.0 degree_Celsius | degC |
| 574694 | 11NPSWRD_WQX-CACO_GULL | 1.5 | deg C | NaN | 1.5 degree_Celsius | degC |
| 574695 | 11NPSWRD_WQX-CACO_GULL | 3.9 | deg C | NaN | 3.9 degree_Celsius | degC |
150773 rows × 6 columns
[34]:
# Examine deg F
temperature_results.loc[df['ResultMeasure/MeasureUnitCode'] == 'deg F']
[34]:
| MonitoringLocationIdentifier | ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Temperature | Units | |
|---|---|---|---|---|---|---|
| 12632 | NALMS-F865245 | 81 | deg F | NaN | 27.222222222222285 degree_Celsius | degF |
| 31004 | 11113300-HOODERD | 59.2 | deg F | NaN | 15.111111111111143 degree_Celsius | degF |
| 31375 | 11113300-HOODERD | 59.1 | deg F | NaN | 15.0555555555556 degree_Celsius | degF |
| 31705 | 11113300-HOODERD | 59.9 | deg F | NaN | 15.500000000000057 degree_Celsius | degF |
| 32253 | 11113300-HOODERD | 56 | deg F | NaN | 13.333333333333371 degree_Celsius | degF |
| ... | ... | ... | ... | ... | ... | ... |
| 560701 | 11NPSWRD_WQX-SAIR_DMF11_HS1 | 56.0 | deg F | NaN | 13.333333333333371 degree_Celsius | degF |
| 560705 | 11NPSWRD_WQX-SAIR_DMF11_OS3 | 62.0 | deg F | NaN | 16.666666666666686 degree_Celsius | degF |
| 560706 | 11NPSWRD_WQX-SAIR_DMF11_OS4 | 61.0 | deg F | NaN | 16.111111111111143 degree_Celsius | degF |
| 560707 | 11NPSWRD_WQX-SAIR_DMF11_OS1 | 59.0 | deg F | NaN | 15.000000000000057 degree_Celsius | degF |
| 560713 | 11NPSWRD_WQX-SAIR_DMF11_S2 | 43.0 | deg F | NaN | 6.111111111111143 degree_Celsius | degF |
695 rows × 6 columns
In the above we can see examples where the results were in deg F and in the result field they’ve been converted into degree_Celsius
[35]:
# Examine missing units
temperature_results.loc[df['ResultMeasure/MeasureUnitCode'].isna()]
[35]:
| MonitoringLocationIdentifier | ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Temperature | Units | |
|---|---|---|---|---|---|---|
| 177453 | AQUINNAH-MEN PND HC | 0 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 0.0 degree_Celsius | degC |
| 177454 | AQUINNAH-MEN PND HC | 0 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 0.0 degree_Celsius | degC |
| 177455 | AQUINNAH-MEN PND HC | 0 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 0.0 degree_Celsius | degC |
| 177456 | AQUINNAH-MEN PND HC | 0 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 0.0 degree_Celsius | degC |
| 177457 | AQUINNAH-MEN PND HC | 0 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 0.0 degree_Celsius | degC |
| ... | ... | ... | ... | ... | ... | ... |
| 263066 | AQUINNAH-MEN PND HC | 0 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 0.0 degree_Celsius | degC |
| 555613 | 11113300-HHPS056 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN | degC |
| 555768 | 11113300-HHPS055 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN | degC |
| 555826 | 11113300-HHPS061 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN | degC |
| 558899 | USGS-414654070002901 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN | degC |
221 rows × 6 columns
We can see where the units were missing, the results were assumed to be in degree_Celsius already
[36]:
# This is also noted in the QA_flag field
list(temperature_results.loc[df['ResultMeasure/MeasureUnitCode'].isna(), 'QA_flag'])[0]
[36]:
'ResultMeasure/MeasureUnitCode: MISSING UNITS, degC assumed'
[37]:
# Look for any without usable results
temperature_results.loc[df['Temperature'].isna()]
[37]:
| MonitoringLocationIdentifier | ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Temperature | Units | |
|---|---|---|---|---|---|---|
| 361 | MERRIMACK_RIVER_WATERSHED_WQX-Hayden-Schofield | 8 | count | NaN | NaN | count |
| 3668 | MERRIMACK_RIVER_WATERSHED_WQX-Manchester | 8 | count | NaN | NaN | count |
| 4193 | MERRIMACK_RIVER_WATERSHED_WQX-Nina-Scarito | 8 | count | NaN | NaN | count |
| 5321 | MERRIMACK_RIVER_WATERSHED_WQX-Misserville | 8 | count | NaN | NaN | count |
| 5731 | MERRIMACK_RIVER_WATERSHED_WQX-Ferrous | 8 | count | NaN | NaN | count |
| ... | ... | ... | ... | ... | ... | ... |
| 555782 | 11113300-HHPS067 | NO DATA | deg C | ResultMeasureValue: "NO DATA" result cannot be... | NaN | degC |
| 555801 | 11113300-HHPS057 | NO DATA | deg C | ResultMeasureValue: "NO DATA" result cannot be... | NaN | degC |
| 555826 | 11113300-HHPS061 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN | degC |
| 555885 | 11113300-HHPS258 | NO DATA | deg C | ResultMeasureValue: "NO DATA" result cannot be... | NaN | degC |
| 558899 | USGS-414654070002901 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN | degC |
226 rows × 6 columns
[38]:
# Aggregate temperature data by station
visualize.station_summary(temperature_results, 'Temperature')
[38]:
| MonitoringLocationIdentifier | cnt | mean | |
|---|---|---|---|
| 0 | 11113300-00-SPB | 21 | 10.909524 |
| 1 | 11113300-00F-KLY | 3 | 20.233333 |
| 2 | 11113300-01-BAK | 10 | 20.600000 |
| 3 | 11113300-01-BVR | 1 | 21.000000 |
| 4 | 11113300-01-CTP | 39 | 24.438462 |
| ... | ... | ... | ... |
| 6506 | WWMD_VA-WWE1149PI | 1 | 18.800000 |
| 6507 | WWMD_VA-WWE1150PI | 1 | 21.300000 |
| 6508 | WWMD_VA-WWE1151PI | 2 | 20.700000 |
| 6509 | WWMD_VA-WWE1152PI | 1 | 20.000000 |
| 6510 | WWMD_VA-WWE1153PI | 1 | 20.800000 |
6511 rows × 3 columns
[39]:
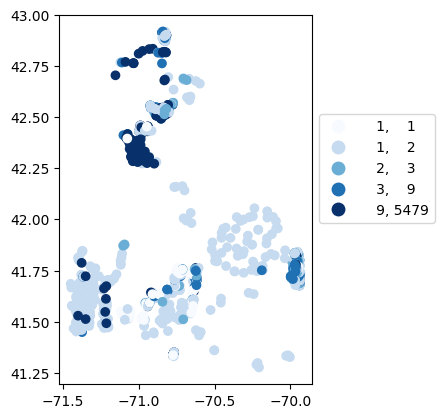
# Map number of usable results at each station
gdf_count = visualize.map_counts(temperature_results, stations_clipped)
gdf_count.plot(column='cnt', cmap='Blues', legend=True, scheme='quantiles', legend_kwds=legend_kwds)
[39]:
<Axes: >
[40]:
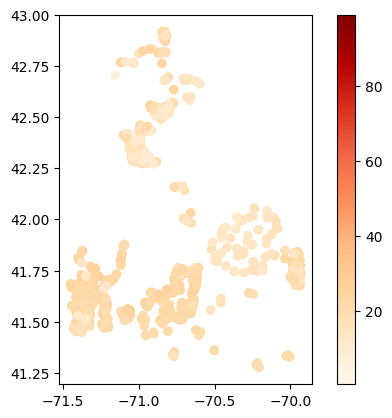
# Map average results at each station
gdf_avg = visualize.map_measure(temperature_results, stations_clipped, 'Temperature')
gdf_avg.plot(column='mean', cmap='OrRd', legend=True)
[40]:
<Axes: >
Dissolved Oxygen (DO)
[41]:
# look at Dissolved oxygen (DO), but this time without intermediate fields
df = harmonize.harmonize(df, 'Dissolved oxygen (DO)')
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(4.6, 'milligram / liter')> <Quantity(9.4, 'milligram / liter')>
<Quantity(10.3, 'milligram / liter')> ...
<Quantity(7.0, 'milligram / liter')> <Quantity(4.0, 'milligram / liter')>
<Quantity(6.01, 'milligram / liter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
Note: Imediately when we run a harmonization function without the intermediate fields they’re deleted.
[42]:
# Look at what was changed
cols = ['MonitoringLocationIdentifier', 'ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'DO']
do_res = df.loc[df['CharacteristicName']=='Dissolved oxygen (DO)', cols]
do_res
[42]:
| MonitoringLocationIdentifier | ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | DO | |
|---|---|---|---|---|---|
| 14 | IRWA-HB | 4.6 | mg/L | NaN | 4.6 milligram / liter |
| 17 | MYRWA-MEB001 | 9.4 | mg/L | NaN | 9.4 milligram / liter |
| 19 | NARS_WQX-MARO-1020 | 10.3 | mg/L | NaN | 10.3 milligram / liter |
| 22 | WTGHA-M46 | 24.7 | mg/L | NaN | 24.7 milligram / liter |
| 23 | MASSDEP-W2412 | 8.7 | mg/L | NaN | 8.7 milligram / liter |
| ... | ... | ... | ... | ... | ... |
| 571625 | IRWA-FB-LL | 6.0 | mg/L | NaN | 6.0 milligram / liter |
| 571626 | HORSLEYWITTEN_WQX-BWB4 | 8.51 | mg/L | NaN | 8.51 milligram / liter |
| 571631 | IRWA-PB05 | 7.0 | mg/L | NaN | 7.0 milligram / liter |
| 571634 | IRWA-PB05 | 4.0 | mg/L | NaN | 4.0 milligram / liter |
| 571635 | HORSLEYWITTEN_WQX-B7 | 6.01 | mg/L | NaN | 6.01 milligram / liter |
94721 rows × 5 columns
[43]:
do_res.loc[do_res['ResultMeasure/MeasureUnitCode']!='mg/l']
[43]:
| MonitoringLocationIdentifier | ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | DO | |
|---|---|---|---|---|---|
| 14 | IRWA-HB | 4.6 | mg/L | NaN | 4.6 milligram / liter |
| 17 | MYRWA-MEB001 | 9.4 | mg/L | NaN | 9.4 milligram / liter |
| 19 | NARS_WQX-MARO-1020 | 10.3 | mg/L | NaN | 10.3 milligram / liter |
| 22 | WTGHA-M46 | 24.7 | mg/L | NaN | 24.7 milligram / liter |
| 23 | MASSDEP-W2412 | 8.7 | mg/L | NaN | 8.7 milligram / liter |
| ... | ... | ... | ... | ... | ... |
| 571625 | IRWA-FB-LL | 6.0 | mg/L | NaN | 6.0 milligram / liter |
| 571626 | HORSLEYWITTEN_WQX-BWB4 | 8.51 | mg/L | NaN | 8.51 milligram / liter |
| 571631 | IRWA-PB05 | 7.0 | mg/L | NaN | 7.0 milligram / liter |
| 571634 | IRWA-PB05 | 4.0 | mg/L | NaN | 4.0 milligram / liter |
| 571635 | HORSLEYWITTEN_WQX-B7 | 6.01 | mg/L | NaN | 6.01 milligram / liter |
93424 rows × 5 columns
Though there were no results in %, the conversion from percent saturation (%) to mg/l is special. This equation is being improved by integrating tempertaure and pressure instead of assuming STP (see DO_saturation())
[44]:
# Aggregate data by station
visualize.station_summary(do_res, 'DO')
[44]:
| MonitoringLocationIdentifier | cnt | mean | |
|---|---|---|---|
| 0 | 11113300-00F-KLY | 3 | 4.040000 |
| 1 | 11113300-01-BAK | 10 | 5.697000 |
| 2 | 11113300-01-BVR | 1 | 6.700000 |
| 3 | 11113300-01-CTP | 37 | 6.945135 |
| 4 | 11113300-01-GOL | 1 | 4.400000 |
| ... | ... | ... | ... |
| 2821 | WWMD_VA-SR5 | 1 | 10.810000 |
| 2822 | WWMD_VA-WF2 | 2 | 6.800000 |
| 2823 | WWMD_VA-WI1 | 1 | 9.510000 |
| 2824 | WWMD_VA-WR2X | 2 | 6.000000 |
| 2825 | WWMD_VA-WR5 | 1 | 8.355000 |
2826 rows × 3 columns
[45]:
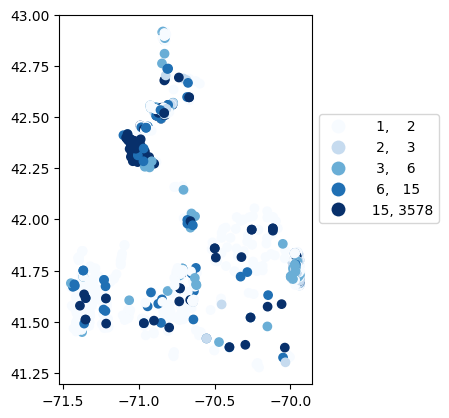
# Map number of usable results at each station
gdf_count = visualize.map_counts(do_res, stations_clipped)
gdf_count.plot(column='cnt', cmap='Blues', legend=True, scheme='quantiles', legend_kwds=legend_kwds)
[45]:
<Axes: >
[46]:
# Map average results at each station
gdf_avg = visualize.map_measure(do_res, stations_clipped, 'DO')
gdf_avg.plot(column='mean', cmap='OrRd', legend=True)
[46]:
<Axes: >
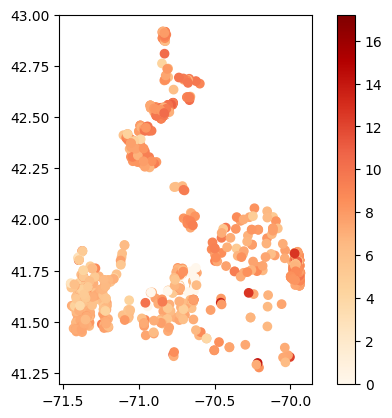
pH
[47]:
# pH, this time looking at a report
df = harmonize.harmonize(df, "pH", errors="ignore", report=True)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'mV' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(7.3, 'dimensionless')> <Quantity(8.16, 'dimensionless')>
<Quantity(7.99, 'dimensionless')> ... <Quantity(6.6, 'dimensionless')>
<Quantity(6.8, 'dimensionless')> <Quantity(7.1, 'dimensionless')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
-Usable results-
count 107704.000000
mean 6.683371
std 4.082578
min -3.124705
25% 5.860000
50% 6.800000
75% 7.670000
max 788.000000
dtype: float64
Unusable results: 4814
Usable results with inferred units: 86150
Results outside threshold (0.0 to 31.17884114461888): 14
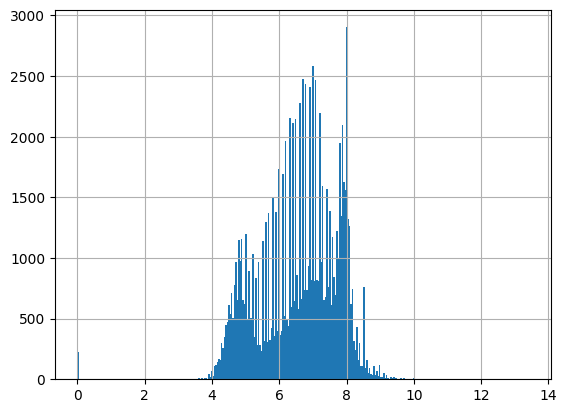
Note the warnings that occur when a unit is not recognized by the package. These occur even when report=False. Future versions could include these as defined units for pH, but here it wouldn’t alter results.
[48]:
df.loc[df['CharacteristicName']=='pH', ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'pH']]
[48]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | pH | |
|---|---|---|---|---|
| 6 | 7.3 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 7.3 dimensionless |
| 9 | 8.16 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 8.16 dimensionless |
| 26 | 7.99 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 7.99 dimensionless |
| 44 | 6.61 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 6.61 dimensionless |
| 46 | 8.23 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 8.23 dimensionless |
| ... | ... | ... | ... | ... |
| 574603 | 6.6 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 6.6 dimensionless |
| 574610 | 6.9 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 6.9 dimensionless |
| 574634 | 6.6 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 6.6 dimensionless |
| 574649 | 6.8 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 6.8 dimensionless |
| 574676 | 7.1 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 7.1 dimensionless |
112518 rows × 4 columns
‘None’ is uninterpretable and replaced with NaN, which then gets replaced with ‘dimensionless’ since pH is unitless
Salinity
[49]:
# Salinity
df = harmonize.harmonize(df, 'Salinity', report=True, errors='ignore')
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/basis.py:154: UserWarning: Mismatched ResultTemperatureBasisText: updated from 25 deg C to @25C (units)
warn(f"Mismatched {flag}", UserWarning)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'deg C' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(30.33, 'Practical_Salinity_Units')>
<Quantity(30.48, 'Practical_Salinity_Units')>
<Quantity(33.3, 'Practical_Salinity_Units')> ...
<Quantity(21.0, 'Practical_Salinity_Units')>
<Quantity(18.7, 'Practical_Salinity_Units')>
<Quantity(10.1, 'Practical_Salinity_Units')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
-Usable results-
count 60420.000000
mean 23.268355
std 20.581826
min -30.000000
25% 13.307500
50% 30.000000
75% 31.500000
max 4003.482834
dtype: float64
Unusable results: 228
Usable results with inferred units: 1
Results outside threshold (0.0 to 146.75931183993742): 3
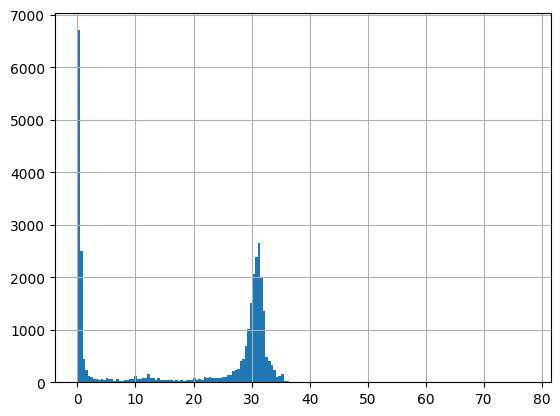
[50]:
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Salinity']
df.loc[df['CharacteristicName']=='Salinity', cols]
[50]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Salinity | |
|---|---|---|---|---|
| 10 | 30.33 | ppt | NaN | 30.33 Practical_Salinity_Units |
| 51 | 30.48 | ppt | NaN | 30.48 Practical_Salinity_Units |
| 56 | 33.3 | ppth | NaN | 33.3 Practical_Salinity_Units |
| 73 | 30.91 | ppt | NaN | 30.91 Practical_Salinity_Units |
| 77 | 0.21 | ppth | NaN | 0.21 Practical_Salinity_Units |
| ... | ... | ... | ... | ... |
| 573337 | 0.4 | PSU | NaN | 0.4 Practical_Salinity_Units |
| 573995 | 4.6 | PSU | NaN | 4.6 Practical_Salinity_Units |
| 574095 | 21.0 | PSU | NaN | 21.0 Practical_Salinity_Units |
| 574108 | 18.7 | PSU | NaN | 18.7 Practical_Salinity_Units |
| 574219 | 10.1 | PSU | NaN | 10.1 Practical_Salinity_Units |
60648 rows × 4 columns
Nitrogen
[51]:
# Nitrogen
df = harmonize.harmonize(df, 'Nitrogen', report=True)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/basis.py:343: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value 'as N' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[mask, basis_col] = basis
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:484: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '['as N' 'as N' 'as N' ... nan nan nan]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
self.df[c_mask] = basis.basis_from_method_spec(self.df[c_mask])
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:395: UserWarning: WARNING: 'cm3/g' UNDEFINED UNIT for Nitrogen
warn("WARNING: " + problem)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/pandas/core/construction.py:627: UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
data = np.asarray(data)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/pandas/core/construction.py:627: UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
data = np.asarray(data)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(0.926976, 'milligram / liter')>
<Quantity(1.571196, 'milligram / liter')>
<Quantity(0.816144, 'milligram / liter')> ...
<Quantity(0.238, 'milligram / liter')>
<Quantity(0.052, 'milligram / liter')>
<Quantity(0.119, 'milligram / liter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/domains.py:277: FutureWarning: Downcasting object dtype arrays on .fillna, .ffill, .bfill is deprecated and will change in a future version. Call result.infer_objects(copy=False) instead. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
sub_df[cols[2]] = sub_df[cols[2]].fillna(sub_df[cols[1]]) # new_fract
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/domains.py:277: FutureWarning: Downcasting object dtype arrays on .fillna, .ffill, .bfill is deprecated and will change in a future version. Call result.infer_objects(copy=False) instead. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
sub_df[cols[2]] = sub_df[cols[2]].fillna(sub_df[cols[1]]) # new_fract
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/domains.py:277: FutureWarning: Downcasting object dtype arrays on .fillna, .ffill, .bfill is deprecated and will change in a future version. Call result.infer_objects(copy=False) instead. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
sub_df[cols[2]] = sub_df[cols[2]].fillna(sub_df[cols[1]]) # new_fract
-Usable results-
count 2508.000000
mean 6.264310
std 104.802798
min 0.000800
25% 0.078000
50% 0.264375
75% 1.030000
max 2800.000000
dtype: float64
Unusable results: 243
Usable results with inferred units: 0
Results outside threshold (0.0 to 635.0810968269224): 6
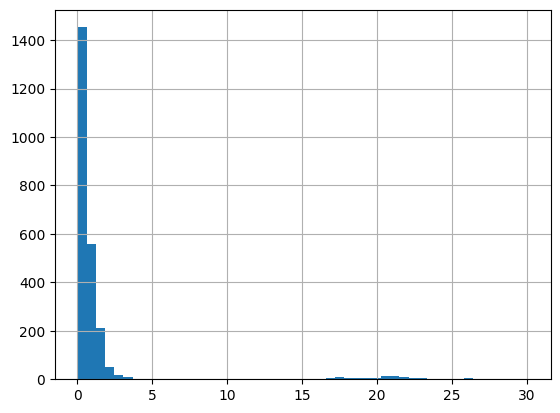
[52]:
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Nitrogen']
df.loc[df['CharacteristicName']=='Nitrogen', cols]
[52]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Nitrogen | |
|---|---|---|---|---|
| 877 | 0.926976 | mg/L | NaN | 0.926976 milligram / liter |
| 953 | 1.571196 | mg/L | NaN | 1.571196 milligram / liter |
| 960 | 0.816144 | mg/L | NaN | 0.816144 milligram / liter |
| 1268 | 0.770448 | mg/L | NaN | 0.770448 milligram / liter |
| 1306 | 0.848832 | mg/L | NaN | 0.848832 milligram / liter |
| ... | ... | ... | ... | ... |
| 574415 | 0.119 | mg/l | NaN | 0.119 milligram / liter |
| 574473 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN |
| 574516 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN |
| 574522 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN |
| 574559 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN |
2751 rows × 4 columns
Conductivity
[53]:
# Conductivity
df = harmonize.harmonize(df, 'Conductivity', report=True, errors='ignore')
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'count' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(590.0, 'microsiemens / centimeter')>
<Quantity(43500.0, 'microsiemens / centimeter')>
<Quantity(349.0, 'microsiemens / centimeter')> ...
<Quantity(45307.0, 'microsiemens / centimeter')>
<Quantity(23758.0, 'microsiemens / centimeter')>
<Quantity(44631.0, 'microsiemens / centimeter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
-Usable results-
count 13515.000000
mean 27165.888787
std 16278.787014
min 0.000000
25% 16570.000000
50% 30700.000000
75% 41833.000000
max 57700.000000
dtype: float64
Unusable results: 82
Usable results with inferred units: 0
Results outside threshold (0.0 to 124838.61087254455): 0
[54]:
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Conductivity']
df.loc[df['CharacteristicName']=='Conductivity', cols]
[54]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Conductivity | |
|---|---|---|---|---|
| 2 | 590 | uS/cm | NaN | 590.0 microsiemens / centimeter |
| 7 | 43.5 | mS/cm | NaN | 43500.0 microsiemens / centimeter |
| 42 | 349 | uS/cm | NaN | 349.0 microsiemens / centimeter |
| 53 | 49.7 | mS/cm | NaN | 49700.0 microsiemens / centimeter |
| 58 | 8 | count | NaN | NaN |
| ... | ... | ... | ... | ... |
| 369376 | 44.393 | mS/cm | NaN | 44393.0 microsiemens / centimeter |
| 369380 | 48.46 | mS/cm | NaN | 48460.0 microsiemens / centimeter |
| 369385 | 45.307 | mS/cm | NaN | 45307.0 microsiemens / centimeter |
| 369388 | 23.758 | mS/cm | NaN | 23758.0 microsiemens / centimeter |
| 369403 | 44.631 | mS/cm | NaN | 44631.0 microsiemens / centimeter |
13597 rows × 4 columns
Chlorophyll a
[55]:
# Chlorophyll a
df = harmonize.harmonize(df, 'Chlorophyll a', report=True)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:395: UserWarning: WARNING: 'ug/cm2' UNDEFINED UNIT for Chlorophyll
warn("WARNING: " + problem)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:395: UserWarning: WARNING: 'ppb' UNDEFINED UNIT for Chlorophyll
warn("WARNING: " + problem)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:395: UserWarning: WARNING: 'ug/m3' UNDEFINED UNIT for Chlorophyll
warn("WARNING: " + problem)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(0.004, 'milligram / liter')>
<Quantity(0.0055, 'milligram / liter')>
<Quantity(0.00948, 'milligram / liter')> ...
<Quantity(0.0007, 'milligram / liter')>
<Quantity(0.0011, 'milligram / liter')>
<Quantity(0.0007, 'milligram / liter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
-Usable results-
count 14059.00000
mean 0.17631
std 1.80612
min -0.00240
25% 0.00180
50% 0.00350
75% 0.00730
max 92.90000
dtype: float64
Unusable results: 204
Usable results with inferred units: 9
Results outside threshold (0.0 to 11.013030176004033): 64
[56]:
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Chlorophyll']
df.loc[df['CharacteristicName']=='Chlorophyll a', cols]
[56]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Chlorophyll | |
|---|---|---|---|---|
| 13 | 4.0 | mg/m3 | NaN | 0.004000000000000001 milligram / liter |
| 288 | 5.50 | ug/L | NaN | 0.0055 milligram / liter |
| 393 | 9.48 | ug/L | NaN | 0.00948 milligram / liter |
| 713 | 3.0 | mg/m3 | NaN | 0.003000000000000001 milligram / liter |
| 1013 | NaN | mg/m3 | ResultMeasureValue: missing (NaN) result | NaN |
| ... | ... | ... | ... | ... |
| 574299 | 0.5 | ug/l | NaN | 0.0005 milligram / liter |
| 574306 | 2.5 | ug/l | NaN | 0.0025 milligram / liter |
| 574319 | 0.7 | ug/l | NaN | 0.0007 milligram / liter |
| 574352 | 1.1 | ug/l | NaN | 0.0011 milligram / liter |
| 574407 | 0.7 | ug/l | NaN | 0.0007 milligram / liter |
14263 rows × 4 columns
Organic Carbon
[57]:
# Organic carbon (%)
df = harmonize.harmonize(df, 'Organic carbon', report=True, errors='ignore')
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'umol/L * H2O' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(7.05, 'milligram / liter')>
<Quantity(7.57, 'milligram / liter')>
<Quantity(3.5, 'milligram / liter')> ...
<Quantity(4.1, 'milligram / liter')>
<Quantity(7.79, 'milligram / liter')>
<Quantity(5.79, 'milligram / liter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
-Usable results-
count 6191.000000
mean 5244.622235
std 35451.442340
min -90000.000000
25% 1.800000
50% 4.900000
75% 8.885000
max 530000.000000
dtype: float64
Unusable results: 89
Usable results with inferred units: 0
Results outside threshold (0.0 to 217953.2762758836): 40
[58]:
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Carbon']
df.loc[df['CharacteristicName']=='Organic carbon', cols]
[58]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Carbon | |
|---|---|---|---|---|
| 1007 | 7.05 | mg/L | NaN | 7.05 milligram / liter |
| 1145 | 7.57 | mg/L | NaN | 7.57 milligram / liter |
| 1199 | 3.5 | mg/L | NaN | 3.5 milligram / liter |
| 3158 | 3.8 | mg/L | NaN | 3.8 milligram / liter |
| 4020 | 3.7 | mg/L | NaN | 3.7 milligram / liter |
| ... | ... | ... | ... | ... |
| 574403 | 7.01 | mg/l | NaN | 7.01 milligram / liter |
| 574472 | 5.96 | mg/l | NaN | 5.96 milligram / liter |
| 574515 | 4.1 | mg/l | NaN | 4.1 milligram / liter |
| 574521 | 7.79 | mg/l | NaN | 7.79 milligram / liter |
| 574558 | 5.79 | mg/l | NaN | 5.79 milligram / liter |
6280 rows × 4 columns
Turbidity (NTU)
[59]:
# Turbidity (NTU)
df = harmonize.harmonize(df, 'Turbidity', report=True, errors='ignore')
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/harmonize.py:149: UserWarning: Bad Turbidity unit: count
warn(f"Bad Turbidity unit: {unit}")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'count' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(7.6, 'Nephelometric_Turbidity_Units')>
<Quantity(1.0, 'Nephelometric_Turbidity_Units')>
<Quantity(4.2, 'Nephelometric_Turbidity_Units')> ...
<Quantity(2.1, 'Nephelometric_Turbidity_Units')>
<Quantity(2.9, 'Nephelometric_Turbidity_Units')>
<Quantity(2.7, 'Nephelometric_Turbidity_Units')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
-Usable results-
count 34461.000000
mean 15.657555
std 72.069369
min -999.000000
25% 1.250000
50% 2.500000
75% 5.770000
max 4100.000000
dtype: float64
Unusable results: 3418
Usable results with inferred units: 275
Results outside threshold (0.0 to 448.07376883334996): 215
[60]:
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Turbidity']
df.loc[df['CharacteristicName']=='Turbidity', cols]
[60]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Turbidity | |
|---|---|---|---|---|
| 0 | 7.6 | NTU | NaN | 7.6 Nephelometric_Turbidity_Units |
| 4 | 1 | NTU | NaN | 1.0 Nephelometric_Turbidity_Units |
| 11 | 4.2 | NTU | NaN | 4.2 Nephelometric_Turbidity_Units |
| 16 | 1.3 | NTU | NaN | 1.3 Nephelometric_Turbidity_Units |
| 21 | 5.2 | NTU | NaN | 5.2 Nephelometric_Turbidity_Units |
| ... | ... | ... | ... | ... |
| 574576 | 1.6 | NTRU | NaN | 1.6 Nephelometric_Turbidity_Units |
| 574586 | 5.8 | NTRU | NaN | 5.8 Nephelometric_Turbidity_Units |
| 574590 | 2.1 | NTRU | NaN | 2.1 Nephelometric_Turbidity_Units |
| 574594 | 2.9 | NTRU | NaN | 2.9 Nephelometric_Turbidity_Units |
| 574598 | 2.7 | NTRU | NaN | 2.7 Nephelometric_Turbidity_Units |
37879 rows × 4 columns
Sediment
[61]:
# Sediment
df = harmonize.harmonize(df, 'Sediment', report=False, errors='ignore')
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'g / H2O' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:134: FutureWarning: The behavior of array concatenation with empty entries is deprecated. In a future version, this will no longer exclude empty items when determining the result dtype. To retain the old behavior, exclude the empty entries before the concat operation.
return pandas.concat(lst_series).sort_index()
[62]:
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Sediment']
df.loc[df['CharacteristicName']=='Sediment', cols]
[62]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Sediment | |
|---|---|---|---|---|
| 521880 | 0.012 | g | NaN | NaN |
| 521881 | 0.0037 | g | NaN | NaN |
| 521882 | 0.0048 | g | NaN | NaN |
| 521883 | 0.001 | g | NaN | NaN |
| 521884 | 0.0088 | g | NaN | NaN |
| ... | ... | ... | ... | ... |
| 573680 | 0.0051 | g | NaN | NaN |
| 573754 | 0.0025 | g | NaN | NaN |
| 573826 | 0.002 | g | NaN | NaN |
| 573865 | 0.0023 | g | NaN | NaN |
| 573911 | 0.0014 | g | NaN | NaN |
4410 rows × 4 columns
Phosphorus
Note: must be merged w/ activities (package runs query by site if not already merged)
[63]:
# Phosphorus
df = harmonize.harmonize(df, 'Phosphorus', errors='ignore')
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/pandas/core/construction.py:627: UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
data = np.asarray(data)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/pandas/core/construction.py:627: UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
data = np.asarray(data)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/pandas/core/construction.py:627: UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
data = np.asarray(data)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'umol/L * H2O' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(0.049, 'milligram / liter')>
<Quantity(0.024, 'milligram / liter')>
<Quantity(0.05, 'milligram / liter')> ...
<Quantity(0.008, 'milligram / liter')>
<Quantity(0.047, 'milligram / liter')>
<Quantity(0.028, 'milligram / liter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
2 Phosphorus sample fractions not in frac_dict
2 Phosphorus sample fractions not in frac_dict found in expected domains, mapped to "Other_Phosphorus"
Note: warnings for unexpected characteristic fractions. Fractions are each seperated out into their own result column.
[64]:
# All Phosphorus
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'TDP_Phosphorus']
df.loc[df['Phosphorus'].notna(), cols]
[64]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | TDP_Phosphorus | |
|---|---|---|---|---|
| 65 | 0.049 | mg/L | NaN | NaN |
| 67 | 0.024 | mg/L | NaN | NaN |
| 72 | 0.05 | mg/L | NaN | NaN |
| 89 | 0.027 | mg/L | NaN | NaN |
| 110 | 0.059712 | mg/L | NaN | NaN |
| ... | ... | ... | ... | ... |
| 573887 | 0.006 | mg/l as P | NaN | 0.006 milligram / liter |
| 573899 | 0.016 | mg/l as P | NaN | NaN |
| 573900 | 0.008 | mg/l as P | NaN | 0.008 milligram / liter |
| 573907 | 0.047 | mg/l as P | NaN | NaN |
| 573908 | 0.028 | mg/l as P | NaN | 0.028 milligram / liter |
19507 rows × 4 columns
[65]:
# Total phosphorus
df.loc[df['TP_Phosphorus'].notna(), cols]
[65]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | TDP_Phosphorus | |
|---|---|---|---|---|
| 65 | 0.049 | mg/L | NaN | NaN |
| 67 | 0.024 | mg/L | NaN | NaN |
| 72 | 0.05 | mg/L | NaN | NaN |
| 89 | 0.027 | mg/L | NaN | NaN |
| 110 | 0.059712 | mg/L | NaN | NaN |
| ... | ... | ... | ... | ... |
| 573861 | 0.036 | mg/l as P | NaN | NaN |
| 573872 | 0.021 | mg/l as P | NaN | NaN |
| 573886 | 0.011 | mg/l as P | NaN | NaN |
| 573899 | 0.016 | mg/l as P | NaN | NaN |
| 573907 | 0.047 | mg/l as P | NaN | NaN |
14234 rows × 4 columns
[66]:
# Total dissolved phosphorus
df.loc[df['TDP_Phosphorus'].notna(), cols]
[66]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | TDP_Phosphorus | |
|---|---|---|---|---|
| 38508 | 0.023 | ppm | NaN | 0.023000000000000003 milligram / liter |
| 43730 | 0.035 | ppm | NaN | 0.03500000000000001 milligram / liter |
| 46336 | 0.017 | ppm | NaN | 0.017000000000000005 milligram / liter |
| 46492 | 0.015 | ppm | NaN | 0.015000000000000003 milligram / liter |
| 48282 | 0.015 | ppm | NaN | 0.015000000000000003 milligram / liter |
| ... | ... | ... | ... | ... |
| 573862 | 0.015 | mg/l as P | NaN | 0.015 milligram / liter |
| 573873 | 0.005 | mg/l as P | NaN | 0.005 milligram / liter |
| 573887 | 0.006 | mg/l as P | NaN | 0.006 milligram / liter |
| 573900 | 0.008 | mg/l as P | NaN | 0.008 milligram / liter |
| 573908 | 0.028 | mg/l as P | NaN | 0.028 milligram / liter |
4857 rows × 4 columns
[67]:
# All other phosphorus sample fractions
df.loc[df['Other_Phosphorus'].notna(), cols]
[67]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | TDP_Phosphorus | |
|---|---|---|---|---|
| 73925 | 0.03525375 | mg/L | NaN | NaN |
| 73992 | 0.107028125 | mg/L | NaN | NaN |
| 74502 | 0.0697675 | mg/L | NaN | NaN |
| 74767 | 0.04356 | mg/L | NaN | NaN |
| 74896 | 0.03654875 | mg/L | NaN | NaN |
| ... | ... | ... | ... | ... |
| 557910 | 2600.0 | mg/kg | NaN | NaN |
| 557948 | 2900.0 | mg/kg | NaN | NaN |
| 558015 | 2500.0 | mg/kg | NaN | NaN |
| 558093 | 2600.0 | mg/kg | NaN | NaN |
| 558102 | 2200.0 | mg/kg | NaN | NaN |
416 rows × 4 columns
Bacteria
Some equivalence assumptions are built-in where bacteria counts that are not equivalent are treated as such because there is no standard way to convert from one to another.
Fecal Coliform
[68]:
# Known unit with bad dimensionality ('Colony_Forming_Units * milliliter')
df = harmonize.harmonize(df, 'Fecal Coliform', report=True, errors='ignore')
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'cfu/100mL' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'MPN/100mL' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'CFU/100mL' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[nan nan nan ... <Quantity(140.0, 'Colony_Forming_Units / milliliter')>
nan <Quantity(1.0, 'Colony_Forming_Units / milliliter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
-Usable results-
count 2462.000000
mean 910.512591
std 6103.365912
min 0.000000
25% 24.000000
50% 93.000000
75% 400.000000
max 250000.000000
dtype: float64
Unusable results: 5811
Usable results with inferred units: 1
Results outside threshold (0.0 to 37530.70806108734): 7
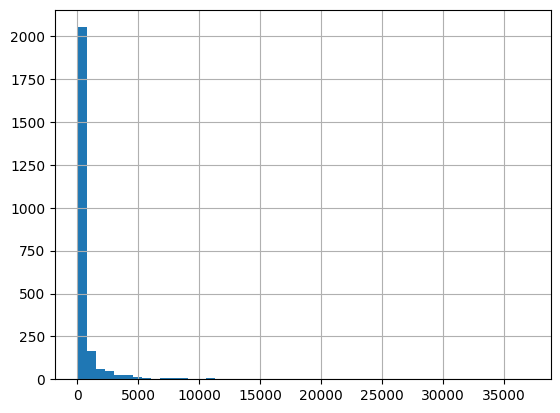
[69]:
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Fecal_Coliform']
df.loc[df['CharacteristicName']=='Fecal Coliform', cols]
[69]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Fecal_Coliform | |
|---|---|---|---|---|
| 22182 | 40 | #/100mL | NaN | NaN |
| 22238 | 9 | #/100mL | NaN | NaN |
| 22258 | 20 | #/100mL | NaN | NaN |
| 22478 | 200 | #/100mL | NaN | NaN |
| 22706 | NO DATA | #/100mL | ResultMeasureValue: "NO DATA" result cannot be... | NaN |
| ... | ... | ... | ... | ... |
| 570290 | 450.0 | cfu/100ml | NaN | 450.0 Colony_Forming_Units / milliliter |
| 570311 | 140.0 | cfu/100ml | NaN | 140.0 Colony_Forming_Units / milliliter |
| 570517 | 180.0 | #/100mL | NaN | NaN |
| 570574 | 1.0 | cfu/100ml | NaN | 1.0 Colony_Forming_Units / milliliter |
| 574601 | NaN | #/100mL | ResultMeasureValue: missing (NaN) result | NaN |
8273 rows × 4 columns
Excherichia Coli
[70]:
# Known unit with bad dimensionality ('Colony_Forming_Units * milliliter')
df = harmonize.harmonize(df, 'Escherichia coli', report=True, errors='ignore')
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:395: UserWarning: WARNING: 'CFUcol/100mL' UNDEFINED UNIT for E_coli
warn("WARNING: " + problem)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'cfu/100mL' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'count' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'MPN/100mL' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: '%' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'CFU/100mL' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[nan nan nan ... <Quantity(44.0, 'Colony_Forming_Units / milliliter')>
<Quantity(14.0, 'Colony_Forming_Units / milliliter')>
<Quantity(390.0, 'Colony_Forming_Units / milliliter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
-Usable results-
count 5190.000000
mean 245.858339
std 1752.146999
min 0.000000
25% 0.500000
50% 30.000000
75% 127.750000
max 72000.000000
dtype: float64
Unusable results: 27668
Usable results with inferred units: 0
Results outside threshold (0.0 to 10758.740332329791): 14
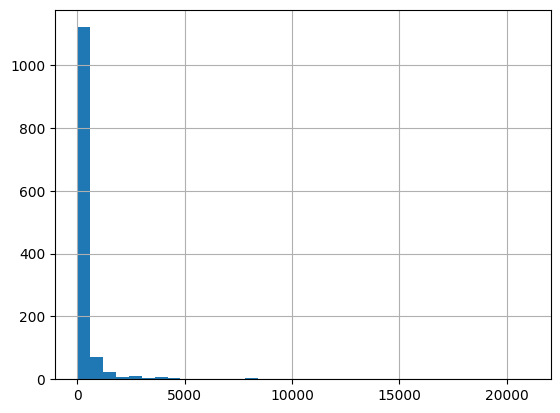
[71]:
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'E_coli']
df.loc[df['CharacteristicName']=='Escherichia coli', cols]
[71]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | E_coli | |
|---|---|---|---|---|
| 15 | 96 | MPN/100mL | NaN | NaN |
| 18 | 58 | #/100mL | NaN | NaN |
| 24 | NaN | MPN/100mL | ResultMeasureValue: missing (NaN) result | NaN |
| 37 | 24200 | MPN/100mL | NaN | NaN |
| 38 | 7270 | MPN/100mL | NaN | NaN |
| ... | ... | ... | ... | ... |
| 574572 | 7.0 | MPN/100 ml | NaN | 7.0 Colony_Forming_Units / milliliter |
| 574585 | 870.0 | MPN/100 ml | NaN | 870.0 Colony_Forming_Units / milliliter |
| 574589 | 44.0 | MPN/100 ml | NaN | 44.0 Colony_Forming_Units / milliliter |
| 574593 | 14.0 | MPN/100 ml | NaN | 14.0 Colony_Forming_Units / milliliter |
| 574597 | 390.0 | MPN/100 ml | NaN | 390.0 Colony_Forming_Units / milliliter |
32858 rows × 4 columns
Combining Salinity and Conductivity
Convert module has various functions to convert from one unit or characteristic to another. Some of these are used within a single characteristic during harmonization (e.g. DO saturation to concentration) while others are intended to model one characteristic as an indicator of another (e.g. estimate salinity from conductivity).
Note: this should only be done after both characteristic fields have been harmonized. Results before and after should be inspected, thresholds for outliers applied, and consider adding a QA_flag for modeled data.
Explore Salinity results:
[72]:
from harmonize_wq import convert
[73]:
# First note initial Salinity info
lst = [x.magnitude for x in list(df['Salinity'].dropna())]
q_sum = sum(lst)
print('Range: {} to {}'.format(min(lst), max(lst)))
print('Results: {} \nMean: {} PSU'.format(len(lst), q_sum/len(lst)))
Range: -30.0 to 4003.4828342857154
Results: 60420
Mean: 23.268354996226737 PSU
[74]:
# Identify extreme outliers
[x for x in lst if x >3200]
[74]:
[4003.4828342857154]
Other fields like units and QA_flag may help understand what caused high values and what results might need to be dropped from consideration
[75]:
# Columns to focus on
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Salinity']
[76]:
# Look at important fields for max 5 values
salinity_series = df['Salinity'][df['Salinity'].notna()]
salinity_series.sort_values(ascending=False, inplace=True)
df[cols][df['Salinity'].isin(salinity_series[0:5])]
[76]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Salinity | |
|---|---|---|---|---|
| 30987 | 804 | ppth | NaN | 804.0 Practical_Salinity_Units |
| 263173 | 70.62 | ppth | NaN | 70.62 Practical_Salinity_Units |
| 263673 | 71.49 | ppth | NaN | 71.49 Practical_Salinity_Units |
| 270683 | 77.6666666666667 | ppt | NaN | 77.6666666666667 Practical_Salinity_Units |
| 564553 | 4980.0 | mg/mL @25C | ResultTemperatureBasisText: updated from 25 de... | 4003.4828342857154 Practical_Salinity_Units |
Detection limits may help understand what caused low values and what results might need to be dropped or updated
[77]:
df = wrangle.add_detection(df, 'Salinity')
cols+=['ResultDetectionConditionText',
'DetectionQuantitationLimitTypeName',
'DetectionQuantitationLimitMeasure/MeasureValue',
'DetectionQuantitationLimitMeasure/MeasureUnitCode']
[78]:
# Look at important fields for min 5 values (often multiple 0.0)
df[cols][df['Salinity'].isin(salinity_series[-5:])]
[78]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Salinity | ResultDetectionConditionText | DetectionQuantitationLimitTypeName | DetectionQuantitationLimitMeasure/MeasureValue | DetectionQuantitationLimitMeasure/MeasureUnitCode | |
|---|---|---|---|---|---|---|---|---|
| 14084 | 0 | ppm | NaN | 0.0 Practical_Salinity_Units | NaN | NaN | NaN | NaN |
| 15918 | 0 | ppm | NaN | 0.0 Practical_Salinity_Units | NaN | NaN | NaN | NaN |
| 17732 | 0 | ppm | NaN | 0.0 Practical_Salinity_Units | NaN | NaN | NaN | NaN |
| 25882 | 0 | ppt | NaN | 0.0 Practical_Salinity_Units | NaN | NaN | NaN | NaN |
| 26524 | 0 | PSS | NaN | 0.0 Practical_Salinity_Units | NaN | NaN | NaN | NaN |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 476555 | 0.0 | ppth | NaN | 0.0 Practical_Salinity_Units | NaN | NaN | NaN | NaN |
| 476611 | 0.0 | ppth | NaN | 0.0 Practical_Salinity_Units | NaN | NaN | NaN | NaN |
| 476657 | 0.0 | ppth | NaN | 0.0 Practical_Salinity_Units | NaN | NaN | NaN | NaN |
| 476703 | 0.0 | ppth | NaN | 0.0 Practical_Salinity_Units | NaN | NaN | NaN | NaN |
| 476715 | 0.0 | ppth | NaN | 0.0 Practical_Salinity_Units | NaN | NaN | NaN | NaN |
806 rows × 8 columns
Explore conductivity results:
[79]:
# Create series for Conductivity values
cond_series = df['Conductivity'].dropna()
cond_series
[79]:
2 590.0 microsiemens / centimeter
7 43500.0 microsiemens / centimeter
42 349.0 microsiemens / centimeter
53 49700.0 microsiemens / centimeter
85 443.0 microsiemens / centimeter
...
369376 44393.0 microsiemens / centimeter
369380 48460.0 microsiemens / centimeter
369385 45307.0 microsiemens / centimeter
369388 23758.0 microsiemens / centimeter
369403 44631.0 microsiemens / centimeter
Name: Conductivity, Length: 13515, dtype: object
Conductivity thresholds from Freshwater Explorer: 10 > x < 5000 us/cm, use a higher threshold for coastal waters
[80]:
# Sort and check other relevant columns before converting (e.g. Salinity)
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Salinity', 'Conductivity']
df.sort_values(by=['Conductivity'], ascending=False, inplace=True)
df.loc[df['Conductivity'].notna(), cols]
[80]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Salinity | Conductivity | |
|---|---|---|---|---|---|
| 279986 | 57700 | uS/cm | NaN | NaN | 57700.0 microsiemens / centimeter |
| 300869 | 52.418 | mS/cm | NaN | NaN | 52418.0 microsiemens / centimeter |
| 318614 | 52.418 | mS/cm | NaN | NaN | 52418.0 microsiemens / centimeter |
| 315665 | 51.618 | mS/cm | NaN | NaN | 51618.0 microsiemens / centimeter |
| 308895 | 51.618 | mS/cm | NaN | NaN | 51618.0 microsiemens / centimeter |
| ... | ... | ... | ... | ... | ... |
| 3992 | 0 | uS/cm | NaN | NaN | 0.0 microsiemens / centimeter |
| 8440 | 0 | uS/cm | NaN | NaN | 0.0 microsiemens / centimeter |
| 71203 | 0 | mS/cm | NaN | NaN | 0.0 microsiemens / centimeter |
| 71050 | 0 | mS/cm | NaN | NaN | 0.0 microsiemens / centimeter |
| 367156 | 0 | uS/cm | NaN | NaN | 0.0 microsiemens / centimeter |
13515 rows × 5 columns
[81]:
# Convert values to PSU and write to Salinity
cond_series = cond_series.apply(str) # Convert to string to convert to dimensionless (PSU)
df.loc[df['Conductivity'].notna(), 'Salinity'] = cond_series.apply(convert.conductivity_to_PSU)
df.loc[df['Conductivity'].notna(), 'Salinity']
[81]:
279986 38.468 dimensionless
300869 34.521 dimensionless
318614 34.521 dimensionless
315665 33.929 dimensionless
308895 33.929 dimensionless
...
3992 0.012 dimensionless
8440 0.012 dimensionless
71203 0.012 dimensionless
71050 0.012 dimensionless
367156 0.012 dimensionless
Name: Salinity, Length: 13515, dtype: object
Datetime
datetime() formats time using dataretrieval and ActivityStart
[82]:
# First inspect the existing unformated fields
cols = ['ActivityStartDate', 'ActivityStartTime/Time', 'ActivityStartTime/TimeZoneCode']
df[cols]
[82]:
| ActivityStartDate | ActivityStartTime/Time | ActivityStartTime/TimeZoneCode | |
|---|---|---|---|
| 279986 | 2021-06-21 | NaN | NaN |
| 300869 | 2022-08-09 | 12:58:00 | EST |
| 318614 | 2022-08-09 | 12:58:00 | EST |
| 315665 | 2022-08-09 | 13:25:00 | EST |
| 308895 | 2022-08-09 | 13:25:00 | EST |
| ... | ... | ... | ... |
| 574706 | 1956-08-16 | NaN | NaN |
| 574707 | 1956-06-16 | NaN | NaN |
| 574708 | 1956-07-01 | NaN | NaN |
| 574709 | 1955-08-16 | NaN | NaN |
| 574710 | 1952-08-16 | NaN | NaN |
574711 rows × 3 columns
[83]:
# 'ActivityStartDate' presserves date where 'Activity_datetime' is NAT due to no time zone
df = clean.datetime(df)
df[['ActivityStartDate', 'Activity_datetime']]
[83]:
| ActivityStartDate | Activity_datetime | |
|---|---|---|
| 279986 | 2021-06-21 | NaT |
| 300869 | 2022-08-09 | 2022-08-09 17:58:00+00:00 |
| 318614 | 2022-08-09 | 2022-08-09 17:58:00+00:00 |
| 315665 | 2022-08-09 | 2022-08-09 18:25:00+00:00 |
| 308895 | 2022-08-09 | 2022-08-09 18:25:00+00:00 |
| ... | ... | ... |
| 574706 | 1956-08-16 | NaT |
| 574707 | 1956-06-16 | NaT |
| 574708 | 1956-07-01 | NaT |
| 574709 | 1955-08-16 | NaT |
| 574710 | 1952-08-16 | NaT |
574711 rows × 2 columns
Activity_datetime combines all three time component columns into UTC. If time is missing this is NaT so a ActivityStartDate column is used to preserve date only.
Depth
Note: Data are often lacking sample depth metadata
[84]:
# Depth of sample (default units='meter')
df = clean.harmonize_depth(df)
#df.loc[df['ResultDepthHeightMeasure/MeasureValue'].dropna(), "Depth"]
df['ResultDepthHeightMeasure/MeasureValue'].dropna()
[84]:
184898 0.15
187034 0.15
118227 0.15
123436 0.15
185567 0.15
...
453100 0.15
453102 0.15
453103 0.15
453104 1.23
453105 1.65
Name: ResultDepthHeightMeasure/MeasureValue, Length: 24059, dtype: float64
Characteristic to Column (long to wide format)
[85]:
# Split single QA column into multiple by characteristic (rename the result to preserve these QA_flags)
df2 = wrangle.split_col(df)
df2
[85]:
| OrganizationIdentifier | OrganizationFormalName | ActivityIdentifier | ActivityStartDate | ActivityStartTime/Time | ActivityStartTime/TimeZoneCode | MonitoringLocationIdentifier | ResultIdentifier | DataLoggerLine | ResultDetectionConditionText | ... | QA_Salinity | QA_pH | QA_Secchi | QA_Fecal_Coliform | QA_Sediment | QA_Conductivity | QA_Carbon | QA_Temperature | QA_Turbidity | QA_Nitrogen | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 279986 | NARS_WQX | EPA National Aquatic Resources Survey (NARS) | NARS_WQX-185450_2021 | 2021-06-21 | NaN | NaN | NARS_WQX-NWC_RI-10033 | STORET-1040684142 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 300869 | WTGHA | Wompanoag Tribe of Gay Head Aquinnah (Tribal) | WTGHA-P3:20220809125800:FM:B | 2022-08-09 | 12:58:00 | EST | WTGHA-P3 | STORET-1041330082 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 318614 | AQUINNAH_WQX | Wampanoag Tribe of Gay Head (Aquinnah) (Tribal) | AQUINNAH_WQX-P3:202208091258:FM | 2022-08-09 | 12:58:00 | EST | AQUINNAH_WQX-P3 | STORET-1057585495 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 315665 | WTGHA | Wompanoag Tribe of Gay Head Aquinnah (Tribal) | WTGHA-P4:20220809132500:FM:B | 2022-08-09 | 13:25:00 | EST | WTGHA-P4 | STORET-1041330098 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 308895 | AQUINNAH_WQX | Wampanoag Tribe of Gay Head (Aquinnah) (Tribal) | AQUINNAH_WQX-P4:202208091325:FM | 2022-08-09 | 13:25:00 | EST | AQUINNAH_WQX-P4 | STORET-1057585502 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 574705 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_DUCK_W_19570701_731710 | 1957-07-01 | NaN | NaN | 11NPSWRD_WQX-CACO_DUCK_W | STORET-740598867 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 574706 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_DUCK_W_19560816_731708 | 1956-08-16 | NaN | NaN | 11NPSWRD_WQX-CACO_DUCK_W | STORET-740598866 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 574707 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_DUCK_W_19560616_731704 | 1956-06-16 | NaN | NaN | 11NPSWRD_WQX-CACO_DUCK_W | STORET-740598862 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 574708 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_DUCK_W_19560701_731705 | 1956-07-01 | NaN | NaN | 11NPSWRD_WQX-CACO_DUCK_W | STORET-740598863 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 574709 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_GREAT_W_19550816_731703 | 1955-08-16 | NaN | NaN | 11NPSWRD_WQX-CACO_GREAT_W | STORET-740649462 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
519723 rows × 118 columns
[86]:
# This expands the single col (QA_flag) out to a number of new columns based on the unique characteristicNames and speciation
print('{} new columns'.format(len(df2.columns) - len(df.columns)))
15 new columns
[87]:
# Note: there are fewer rows because NAN results are also dropped in this step
print('{} fewer rows'.format(len(df)-len(df2)))
54988 fewer rows
[88]:
#Examine Carbon flags from earlier in notebook (note these are empty now because NAN is dropped)
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'Carbon', 'QA_Carbon']
df2.loc[df2['QA_Carbon'].notna(), cols]
[88]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | Carbon | QA_Carbon |
|---|
Next the table is divided into the columns of interest (main_df) and characteristic specific metadata (chars_df)
[89]:
# split table into main and characteristics tables
main_df, chars_df = wrangle.split_table(df2)
[90]:
# Columns still in main table
main_df.columns
[90]:
Index(['OrganizationIdentifier', 'OrganizationFormalName',
'ActivityIdentifier', 'MonitoringLocationIdentifier', 'ProviderName',
'Secchi', 'Temperature', 'DO', 'pH', 'Salinity', 'Nitrogen',
'Speciation', 'TOTAL NITROGEN_ MIXED FORMS', 'Conductivity',
'Chlorophyll', 'Carbon', 'Turbidity', 'Sediment', 'Phosphorus',
'TP_Phosphorus', 'TDP_Phosphorus', 'Other_Phosphorus', 'Fecal_Coliform',
'E_coli', 'DetectionQuantitationLimitTypeName',
'DetectionQuantitationLimitMeasure/MeasureValue',
'DetectionQuantitationLimitMeasure/MeasureUnitCode',
'Activity_datetime', 'Depth', 'QA_TP_Phosphorus', 'QA_TDP_Phosphorus',
'QA_Other_Phosphorus', 'QA_DO', 'QA_E_coli', 'QA_Chlorophyll',
'QA_Salinity', 'QA_pH', 'QA_Secchi', 'QA_Fecal_Coliform', 'QA_Sediment',
'QA_Conductivity', 'QA_Carbon', 'QA_Temperature', 'QA_Turbidity',
'QA_Nitrogen'],
dtype='object')
[91]:
# look at main table results (first 5)
main_df.head()
[91]:
| OrganizationIdentifier | OrganizationFormalName | ActivityIdentifier | MonitoringLocationIdentifier | ProviderName | Secchi | Temperature | DO | pH | Salinity | ... | QA_Salinity | QA_pH | QA_Secchi | QA_Fecal_Coliform | QA_Sediment | QA_Conductivity | QA_Carbon | QA_Temperature | QA_Turbidity | QA_Nitrogen | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 279986 | NARS_WQX | EPA National Aquatic Resources Survey (NARS) | NARS_WQX-185450_2021 | NARS_WQX-NWC_RI-10033 | STORET | NaN | NaN | NaN | NaN | 38.468 dimensionless | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 300869 | WTGHA | Wompanoag Tribe of Gay Head Aquinnah (Tribal) | WTGHA-P3:20220809125800:FM:B | WTGHA-P3 | STORET | NaN | NaN | NaN | NaN | 34.521 dimensionless | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 318614 | AQUINNAH_WQX | Wampanoag Tribe of Gay Head (Aquinnah) (Tribal) | AQUINNAH_WQX-P3:202208091258:FM | AQUINNAH_WQX-P3 | STORET | NaN | NaN | NaN | NaN | 34.521 dimensionless | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 315665 | WTGHA | Wompanoag Tribe of Gay Head Aquinnah (Tribal) | WTGHA-P4:20220809132500:FM:B | WTGHA-P4 | STORET | NaN | NaN | NaN | NaN | 33.929 dimensionless | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 308895 | AQUINNAH_WQX | Wampanoag Tribe of Gay Head (Aquinnah) (Tribal) | AQUINNAH_WQX-P4:202208091325:FM | AQUINNAH_WQX-P4 | STORET | NaN | NaN | NaN | NaN | 33.929 dimensionless | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
5 rows × 45 columns
[92]:
# Empty columns that could be dropped (Mostly QA columns)
cols = list(main_df.columns)
x = main_df.dropna(axis=1, how='all')
[col for col in cols if col not in x.columns]
[92]:
['Sediment',
'QA_TDP_Phosphorus',
'QA_DO',
'QA_Sediment',
'QA_Conductivity',
'QA_Carbon']
[93]:
# Map average results at each station
gdf_avg = visualize.map_measure(main_df, stations_clipped, 'Temperature')
gdf_avg.plot(column='mean', cmap='OrRd', legend=True)
[93]:
<Axes: >