Cape Cod - Simple workflow
Standardize, clean and wrangle Water Quality Portal data in Cape Cod into more analytic-ready formats using the harmonize_wq package
US EPA’s Water Quality Portal (WQP) aggregates water quality, biological, and physical data provided by many organizations and has become an essential resource with tools to query and retrieval data using python or R. Given the variety of data and variety of data originators, using the data in analysis often requires data cleaning to ensure it meets the required quality standards and data wrangling to get it in a more analytic-ready format. Recognizing the definition of analysis-ready varies depending on the analysis, the harmonixe_wq package is intended to be a flexible water quality specific framework to help:
Identify differences in data units (including speciation and basis)
Identify differences in sampling or analytic methods
Resolve data errors using transparent assumptions
Reduce data to the columns that are most commonly needed
Transform data from long to wide format
Domain experts must decide what data meets their quality standards for data comparability and any thresholds for acceptance or rejection.
Simple workflow
This example steps through a typical workflow in Cape Cod to demonstrate commonly used functionality
Install the required libraries
[1]:
import sys
#!python -m pip uninstall harmonize-wq --yes
# Use pip to install the package from pypi or the latest from github
#!{sys.executable} -m pip install harmonize-wq
# For latest dev version
#!{sys.executable} -m pip install git+https://github.com/USEPA/harmonize-wq.git@new_release_0-3-8
Create dataretrieval query using a polygon for Area Of Interest
[2]:
from harmonize_wq import wrangle
[3]:
# Read geometry for Area of Interest from geojson file url
# NOTE: alternatively you can direct it to a local shapefile
aoi_url = 'https://github.com/jbousquin/test_notebook/raw/main/temperature_data/NewEngland.geojson'
[4]:
# Map aoi (geojson is WGS1984 standard)
wrangle.as_gdf(aoi_url).plot()
[4]:
<Axes: >
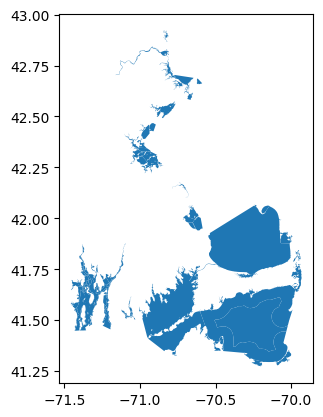
The Area Of Interest is made up of many small polygons. The query will be built from the total extent of these polygons, but alternatively could be restricted to the bounding box for just one such polygon.
[5]:
# Bounding box string for total extent
bBox = wrangle.get_bounding_box(aoi_url)
# Bounding box string for specific polygon by index
#bBox = wrangle.get_bounding_box(aoi_url, idx=0)
[6]:
# Build query
query = {'characteristicName': ['Phosphorus',
'Temperature, water',
'Depth, Secchi disk depth',
'Dissolved oxygen (DO)',
'Salinity',
'pH',
'Nitrogen',
'Conductivity',
'Organic carbon',
'Chlorophyll a',
'Turbidity',
'Sediment',
'Fecal Coliform',
'Escherichia coli']}
query['bBox'] = bBox
query['dataProfile'] = 'narrowResult'
Retrieve data
Stations will be retrieved using the query criteria seperately after results. md_narrow is metadata documenting the query not used here but meant for reproducibility
[7]:
import dataretrieval.wqp as wqp
[8]:
# Query for results
# Note: large quieries like this can take up a lot of RAM and may give a DtypeWarning,
# set low_memory=False or provide dataTypes for columns to use less memory.
res_narrow, md_narrow = wqp.get_results(**query)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/dataretrieval/wqp.py:153: DtypeWarning: Columns (8,10,13,15,17,19,20,21,22,23,28,31,33,34,36,38,60,63,64,65,66,67,68,69,70,71,72) have mixed types. Specify dtype option on import or set low_memory=False.
df = pd.read_csv(StringIO(response.text), delimiter=",")
[9]:
# Look at initial results (input)
df = res_narrow
df
[9]:
| OrganizationIdentifier | OrganizationFormalName | ActivityIdentifier | ActivityStartDate | ActivityStartTime/Time | ActivityStartTime/TimeZoneCode | MonitoringLocationIdentifier | ResultIdentifier | DataLoggerLine | ResultDetectionConditionText | ... | AnalysisEndTime/TimeZoneCode | ResultLaboratoryCommentCode | ResultLaboratoryCommentText | ResultDetectionQuantitationLimitUrl | LaboratoryAccreditationIndicator | LaboratoryAccreditationAuthorityName | TaxonomistAccreditationIndicator | TaxonomistAccreditationAuthorityName | LabSamplePreparationUrl | ProviderName | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | BRC | Blackstone River Coalition (Volunteer) | BRC-C-02-02-020:20131012040800:FM:0.2083333333... | 2013-10-12 | 04:08:00 | EST | BRC-C-02-02-020 | STORET-1039097035 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | STORET |
| 1 | OARS | OARS - For the Assabet Sudbury & Concord River... | OARS-CND-161:20130818:0651:FM:1 | 2013-08-18 | 06:51:00 | EDT | OARS-CND-161 | STORET-838568413 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | STORET |
| 2 | OARS | OARS - For the Assabet Sudbury & Concord River... | OARS-SUD-064:20130721:0610:FM:1 | 2013-07-21 | 06:10:00 | EDT | OARS-SUD-064 | STORET-838568309 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | STORET |
| 3 | CRWA | Charles River Watershed Association (Massachus... | CRWA-CYN20130809ROBTemp01 | 2013-08-09 | 11:14:33 | EST | CRWA-ROB | STORET-591631481 | 130809111433.0 | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | STORET |
| 4 | WTGHA | Wompanoag Tribe of Gay Head Aquinnah (Tribal) | WTGHA-m41:20131017:FM:B | 2013-10-17 | NaN | NaN | WTGHA-M41 | STORET-1041401688 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | STORET |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 574706 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_DUCK_W_19560816_731708 | 1956-08-16 | NaN | NaN | 11NPSWRD_WQX-CACO_DUCK_W | STORET-740598866 | NaN | NaN | ... | NaN | NaN | NaN | https://www.waterqualitydata.us/data/providers... | NaN | NaN | NaN | NaN | NaN | STORET |
| 574707 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_DUCK_W_19560616_731704 | 1956-06-16 | NaN | NaN | 11NPSWRD_WQX-CACO_DUCK_W | STORET-740598862 | NaN | NaN | ... | NaN | NaN | NaN | https://www.waterqualitydata.us/data/providers... | NaN | NaN | NaN | NaN | NaN | STORET |
| 574708 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_DUCK_W_19560701_731705 | 1956-07-01 | NaN | NaN | 11NPSWRD_WQX-CACO_DUCK_W | STORET-740598863 | NaN | NaN | ... | NaN | NaN | NaN | https://www.waterqualitydata.us/data/providers... | NaN | NaN | NaN | NaN | NaN | STORET |
| 574709 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_GREAT_W_19550816_731703 | 1955-08-16 | NaN | NaN | 11NPSWRD_WQX-CACO_GREAT_W | STORET-740649462 | NaN | NaN | ... | NaN | NaN | NaN | https://www.waterqualitydata.us/data/providers... | NaN | NaN | NaN | NaN | NaN | STORET |
| 574710 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_SLOUGH_19520816_731702 | 1952-08-16 | NaN | NaN | 11NPSWRD_WQX-CACO_SLOUGH | STORET-740745800 | NaN | Not Detected | ... | NaN | NaN | NaN | https://www.waterqualitydata.us/data/providers... | NaN | NaN | NaN | NaN | NaN | STORET |
574711 rows × 78 columns
Harmonize data using defaults
[10]:
from harmonize_wq import harmonize
from harmonize_wq import location
from harmonize_wq import visualize
[11]:
# Harmonize it and look at how it changed (output)
# Note: 'ignore' unit dimensionality errors will warn when they are encountered and replace with NaN
# Note: depending on the Pint version this may cause a UnitStrippedWarning
df_harmonized = harmonize.harmonize_all(df, errors='ignore')
df_harmonized
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/clean.py:360: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value 'ResultMeasureValue: missing (NaN) result' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[mask & (df_out["QA_flag"].isna()), "QA_flag"] = flag
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:395: UserWarning: WARNING: 'ug/cm2' UNDEFINED UNIT for Chlorophyll
warn("WARNING: " + problem)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:395: UserWarning: WARNING: 'ppb' UNDEFINED UNIT for Chlorophyll
warn("WARNING: " + problem)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:395: UserWarning: WARNING: 'ug/m3' UNDEFINED UNIT for Chlorophyll
warn("WARNING: " + problem)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(0.004, 'milligram / liter')>
<Quantity(0.0055, 'milligram / liter')>
<Quantity(0.00948, 'milligram / liter')> ...
<Quantity(0.0007, 'milligram / liter')>
<Quantity(0.0011, 'milligram / liter')>
<Quantity(0.0007, 'milligram / liter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'count' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(590.0, 'microsiemens / centimeter')>
<Quantity(43500.0, 'microsiemens / centimeter')>
<Quantity(349.0, 'microsiemens / centimeter')> ...
<Quantity(45307.0, 'microsiemens / centimeter')>
<Quantity(23758.0, 'microsiemens / centimeter')>
<Quantity(44631.0, 'microsiemens / centimeter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: '%' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(3.25, 'meter')> <Quantity(4.25, 'meter')>
<Quantity(3.5, 'meter')> ... <Quantity(17.0, 'meter')>
<Quantity(16.0, 'meter')> <Quantity(7.8, 'meter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(4.6, 'milligram / liter')> <Quantity(9.4, 'milligram / liter')>
<Quantity(10.3, 'milligram / liter')> ...
<Quantity(7.0, 'milligram / liter')> <Quantity(4.0, 'milligram / liter')>
<Quantity(6.01, 'milligram / liter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:395: UserWarning: WARNING: 'CFUcol/100mL' UNDEFINED UNIT for E_coli
warn("WARNING: " + problem)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'cfu/100mL' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: '%' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'CFU/100mL' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'MPN/100mL' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'count' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[nan nan nan ... <Quantity(44.0, 'Colony_Forming_Units / milliliter')>
<Quantity(14.0, 'Colony_Forming_Units / milliliter')>
<Quantity(390.0, 'Colony_Forming_Units / milliliter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'cfu/100mL' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'MPN/100mL' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'CFU/100mL' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[nan nan nan ... <Quantity(140.0, 'Colony_Forming_Units / milliliter')>
nan <Quantity(1.0, 'Colony_Forming_Units / milliliter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/basis.py:343: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value 'as N' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[mask, basis_col] = basis
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:484: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '['as N' 'as N' 'as N' ... nan nan nan]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
self.df[c_mask] = basis.basis_from_method_spec(self.df[c_mask])
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:395: UserWarning: WARNING: 'cm3/g' UNDEFINED UNIT for Nitrogen
warn("WARNING: " + problem)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/pandas/core/construction.py:627: UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
data = np.asarray(data)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/pandas/core/construction.py:627: UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
data = np.asarray(data)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(0.926976, 'milligram / liter')>
<Quantity(1.571196, 'milligram / liter')>
<Quantity(0.816144, 'milligram / liter')> ...
<Quantity(0.238, 'milligram / liter')>
<Quantity(0.052, 'milligram / liter')>
<Quantity(0.119, 'milligram / liter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/domains.py:277: FutureWarning: Downcasting object dtype arrays on .fillna, .ffill, .bfill is deprecated and will change in a future version. Call result.infer_objects(copy=False) instead. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
sub_df[cols[2]] = sub_df[cols[2]].fillna(sub_df[cols[1]]) # new_fract
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/domains.py:277: FutureWarning: Downcasting object dtype arrays on .fillna, .ffill, .bfill is deprecated and will change in a future version. Call result.infer_objects(copy=False) instead. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
sub_df[cols[2]] = sub_df[cols[2]].fillna(sub_df[cols[1]]) # new_fract
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/domains.py:277: FutureWarning: Downcasting object dtype arrays on .fillna, .ffill, .bfill is deprecated and will change in a future version. Call result.infer_objects(copy=False) instead. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
sub_df[cols[2]] = sub_df[cols[2]].fillna(sub_df[cols[1]]) # new_fract
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'umol/L * H2O' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(7.05, 'milligram / liter')>
<Quantity(7.57, 'milligram / liter')>
<Quantity(3.5, 'milligram / liter')> ...
<Quantity(4.1, 'milligram / liter')>
<Quantity(7.79, 'milligram / liter')>
<Quantity(5.79, 'milligram / liter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/pandas/core/construction.py:627: UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
data = np.asarray(data)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/pandas/core/construction.py:627: UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
data = np.asarray(data)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/pandas/core/construction.py:627: UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
data = np.asarray(data)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'umol/L * H2O' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(0.049, 'milligram / liter')>
<Quantity(0.024, 'milligram / liter')>
<Quantity(0.05, 'milligram / liter')> ...
<Quantity(0.008, 'milligram / liter')>
<Quantity(0.047, 'milligram / liter')>
<Quantity(0.028, 'milligram / liter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
2 Phosphorus sample fractions not in frac_dict
2 Phosphorus sample fractions not in frac_dict found in expected domains, mapped to "Other_Phosphorus"
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/basis.py:154: UserWarning: Mismatched ResultTemperatureBasisText: updated from 25 deg C to @25C (units)
warn(f"Mismatched {flag}", UserWarning)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'deg C' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(30.33, 'Practical_Salinity_Units')>
<Quantity(30.48, 'Practical_Salinity_Units')>
<Quantity(33.3, 'Practical_Salinity_Units')> ...
<Quantity(21.0, 'Practical_Salinity_Units')>
<Quantity(18.7, 'Practical_Salinity_Units')>
<Quantity(10.1, 'Practical_Salinity_Units')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'g / H2O' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:134: FutureWarning: The behavior of array concatenation with empty entries is deprecated. In a future version, this will no longer exclude empty items when determining the result dtype. To retain the old behavior, exclude the empty entries before the concat operation.
return pandas.concat(lst_series).sort_index()
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'count' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(22.18, 'degree_Celsius')> <Quantity(23.01, 'degree_Celsius')>
<Quantity(17.39, 'degree_Celsius')> ... <Quantity(4.0, 'degree_Celsius')>
<Quantity(1.5, 'degree_Celsius')> <Quantity(3.9, 'degree_Celsius')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/harmonize.py:149: UserWarning: Bad Turbidity unit: count
warn(f"Bad Turbidity unit: {unit}")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'count' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(7.6, 'Nephelometric_Turbidity_Units')>
<Quantity(1.0, 'Nephelometric_Turbidity_Units')>
<Quantity(4.2, 'Nephelometric_Turbidity_Units')> ...
<Quantity(2.1, 'Nephelometric_Turbidity_Units')>
<Quantity(2.9, 'Nephelometric_Turbidity_Units')>
<Quantity(2.7, 'Nephelometric_Turbidity_Units')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'mV' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(7.3, 'dimensionless')> <Quantity(8.16, 'dimensionless')>
<Quantity(7.99, 'dimensionless')> ... <Quantity(6.6, 'dimensionless')>
<Quantity(6.8, 'dimensionless')> <Quantity(7.1, 'dimensionless')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
[11]:
| OrganizationIdentifier | OrganizationFormalName | ActivityIdentifier | ActivityStartDate | ActivityStartTime/Time | ActivityStartTime/TimeZoneCode | MonitoringLocationIdentifier | ResultIdentifier | DataLoggerLine | ResultDetectionConditionText | ... | Carbon | Phosphorus | TP_Phosphorus | TDP_Phosphorus | Other_Phosphorus | Salinity | Sediment | Temperature | Turbidity | pH | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | BRC | Blackstone River Coalition (Volunteer) | BRC-C-02-02-020:20131012040800:FM:0.2083333333... | 2013-10-12 | 04:08:00 | EST | BRC-C-02-02-020 | STORET-1039097035 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 7.6 Nephelometric_Turbidity_Units | NaN |
| 1 | OARS | OARS - For the Assabet Sudbury & Concord River... | OARS-CND-161:20130818:0651:FM:1 | 2013-08-18 | 06:51:00 | EDT | OARS-CND-161 | STORET-838568413 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 22.18 degree_Celsius | NaN | NaN |
| 2 | OARS | OARS - For the Assabet Sudbury & Concord River... | OARS-SUD-064:20130721:0610:FM:1 | 2013-07-21 | 06:10:00 | EDT | OARS-SUD-064 | STORET-838568309 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 3 | CRWA | Charles River Watershed Association (Massachus... | CRWA-CYN20130809ROBTemp01 | 2013-08-09 | 11:14:33 | EST | CRWA-ROB | STORET-591631481 | 130809111433.0 | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 23.01 degree_Celsius | NaN | NaN |
| 4 | WTGHA | Wompanoag Tribe of Gay Head Aquinnah (Tribal) | WTGHA-m41:20131017:FM:B | 2013-10-17 | NaN | NaN | WTGHA-M41 | STORET-1041401688 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 1.0 Nephelometric_Turbidity_Units | NaN |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 574706 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_DUCK_W_19560816_731708 | 1956-08-16 | NaN | NaN | 11NPSWRD_WQX-CACO_DUCK_W | STORET-740598866 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 574707 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_DUCK_W_19560616_731704 | 1956-06-16 | NaN | NaN | 11NPSWRD_WQX-CACO_DUCK_W | STORET-740598862 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 574708 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_DUCK_W_19560701_731705 | 1956-07-01 | NaN | NaN | 11NPSWRD_WQX-CACO_DUCK_W | STORET-740598863 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 574709 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_GREAT_W_19550816_731703 | 1955-08-16 | NaN | NaN | 11NPSWRD_WQX-CACO_GREAT_W | STORET-740649462 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 574710 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_SLOUGH_19520816_731702 | 1952-08-16 | NaN | NaN | 11NPSWRD_WQX-CACO_SLOUGH | STORET-740745800 | NaN | Not Detected | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
574711 rows × 98 columns
[12]:
# Get harmonized stations clipped to the Area of Interest
stations_gdf, stations, site_md = location.get_harmonized_stations(query, aoi=aoi_url)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/clean.py:356: FutureWarning: Logical ops (and, or, xor) between Pandas objects and dtype-less sequences (e.g. list, tuple) are deprecated and will raise in a future version. Wrap the object in a Series, Index, or np.array before operating instead.
cond_notna = mask & (df_out["QA_flag"].notna()) # Mask cond and not NA
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/clean.py:360: FutureWarning: Logical ops (and, or, xor) between Pandas objects and dtype-less sequences (e.g. list, tuple) are deprecated and will raise in a future version. Wrap the object in a Series, Index, or np.array before operating instead.
df_out.loc[mask & (df_out["QA_flag"].isna()), "QA_flag"] = flag
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/clean.py:360: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value 'LatitudeMeasure: Imprecise: lessthan3decimaldigits' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[mask & (df_out["QA_flag"].isna()), "QA_flag"] = flag
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/clean.py:356: FutureWarning: Logical ops (and, or, xor) between Pandas objects and dtype-less sequences (e.g. list, tuple) are deprecated and will raise in a future version. Wrap the object in a Series, Index, or np.array before operating instead.
cond_notna = mask & (df_out["QA_flag"].notna()) # Mask cond and not NA
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/clean.py:360: FutureWarning: Logical ops (and, or, xor) between Pandas objects and dtype-less sequences (e.g. list, tuple) are deprecated and will raise in a future version. Wrap the object in a Series, Index, or np.array before operating instead.
df_out.loc[mask & (df_out["QA_flag"].isna()), "QA_flag"] = flag
[13]:
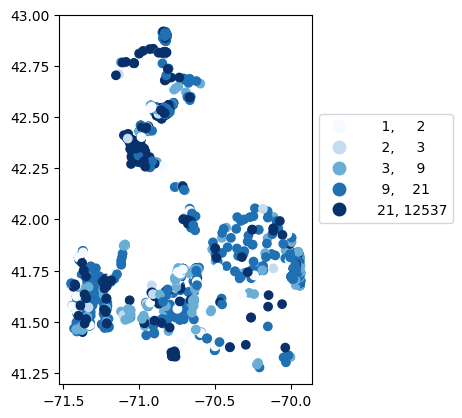
# Map number of usable results at each station
gdf_count = visualize.map_counts(df_harmonized, stations_gdf)
legend_kwds={"fmt": "{:.0f}", 'bbox_to_anchor':(1, 0.75)}
gdf_count.plot(column='cnt', cmap='Blues', legend=True, scheme='quantiles', legend_kwds=legend_kwds)
[13]:
<Axes: >
[14]:
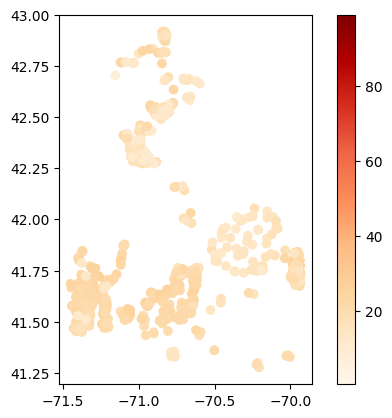
# Map average temperature results at each station
gdf_temperature = visualize.map_measure(df_harmonized, stations_gdf, 'Temperature')
gdf_temperature.plot(column='mean', cmap='OrRd', legend=True)
[14]:
<Axes: >
Clean additional columns of data
[15]:
from harmonize_wq import clean
[16]:
df_cleaned = clean.datetime(df_harmonized) # datetime
df_cleaned = clean.harmonize_depth(df_cleaned) # Sample depth
Reduce data to the columns that are most commonly needed
There are many columns in the dataframe that are characteristic specific, that is they have different values for the same sample depending on the characteristic. To ensure one result for each sample after the transformation of the data these columns must either be split, generating a new column for each characteristic with values, or moved out from the table if not being used.
[17]:
# Split single QA column into multiple by characteristic (rename the result to preserve these QA_flags)
df_expanded = wrangle.split_col(df_cleaned)
df_expanded
[17]:
| OrganizationIdentifier | OrganizationFormalName | ActivityIdentifier | ActivityStartDate | ActivityStartTime/Time | ActivityStartTime/TimeZoneCode | MonitoringLocationIdentifier | ResultIdentifier | DataLoggerLine | ResultDetectionConditionText | ... | QA_Salinity | QA_E_coli | QA_DO | QA_Nitrogen | QA_Chlorophyll | QA_Secchi | QA_pH | QA_Temperature | QA_Conductivity | QA_Carbon | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | BRC | Blackstone River Coalition (Volunteer) | BRC-C-02-02-020:20131012040800:FM:0.2083333333... | 2013-10-12 | 04:08:00 | EST | BRC-C-02-02-020 | STORET-1039097035 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 1 | OARS | OARS - For the Assabet Sudbury & Concord River... | OARS-CND-161:20130818:0651:FM:1 | 2013-08-18 | 06:51:00 | EDT | OARS-CND-161 | STORET-838568413 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 2 | OARS | OARS - For the Assabet Sudbury & Concord River... | OARS-SUD-064:20130721:0610:FM:1 | 2013-07-21 | 06:10:00 | EDT | OARS-SUD-064 | STORET-838568309 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 3 | CRWA | Charles River Watershed Association (Massachus... | CRWA-CYN20130809ROBTemp01 | 2013-08-09 | 11:14:33 | EST | CRWA-ROB | STORET-591631481 | 130809111433.0 | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 4 | WTGHA | Wompanoag Tribe of Gay Head Aquinnah (Tribal) | WTGHA-m41:20131017:FM:B | 2013-10-17 | NaN | NaN | WTGHA-M41 | STORET-1041401688 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 574705 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_DUCK_W_19570701_731710 | 1957-07-01 | NaN | NaN | 11NPSWRD_WQX-CACO_DUCK_W | STORET-740598867 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 574706 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_DUCK_W_19560816_731708 | 1956-08-16 | NaN | NaN | 11NPSWRD_WQX-CACO_DUCK_W | STORET-740598866 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 574707 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_DUCK_W_19560616_731704 | 1956-06-16 | NaN | NaN | 11NPSWRD_WQX-CACO_DUCK_W | STORET-740598862 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 574708 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_DUCK_W_19560701_731705 | 1956-07-01 | NaN | NaN | 11NPSWRD_WQX-CACO_DUCK_W | STORET-740598863 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 574709 | 11NPSWRD_WQX | National Park Service Water Resources Division | 11NPSWRD_WQX-CACO_GREAT_W_19550816_731703 | 1955-08-16 | NaN | NaN | 11NPSWRD_WQX-CACO_GREAT_W | STORET-740649462 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
519723 rows × 115 columns
[18]:
# Split table into main with columns of interest and characteristic specific columns/metadata
main_df, chars_df = wrangle.split_table(df_expanded)
[19]:
# Drop empty columns (QA columns without flags)
main_df_small = main_df.dropna(axis=1, how='all')
Transform data from long to wide format
[20]:
transformed_df = wrangle.collapse_results(main_df_small)
Results are collapsed by retaining the first result that isn’t NAN. There can be several reasons for multiple results for the same parameter/characteristic sampled at the same station, time and by the same organization. The collapse_results function assumes the user has already reviewed the quality of all results and narrowed down instances of multiple results to only the desired/best/highest quality result before running this function.