Pensacola Bay FL - Detailed step-by-step
Standardize, clean and wrangle Water Quality Portal data in Pensacola and Perdido Bays into more analytic-ready formats using the harmonize_wq package
US EPA’s Water Quality Portal (WQP) aggregates water quality, biological, and physical data provided by many organizations and has become an essential resource with tools to query and retrieval data using python or R. Given the variety of data and variety of data originators, using the data in analysis often requires data cleaning to ensure it meets the required quality standards and data wrangling to get it in a more analytic-ready format. Recognizing the definition of analysis-ready varies depending on the analysis, the harmonixe_wq package is intended to be a flexible water quality specific framework to help:
Identify differences in data units (including speciation and basis)
Identify differences in sampling or analytic methods
Resolve data errors using transparent assumptions
Reduce data to the columns that are most commonly needed
Transform data from long to wide format
Domain experts must decide what data meets their quality standards for data comparability and any thresholds for acceptance or rejection.
Detailed step-by-step workflow
This example workflow takes a deeper dive into some of the expanded functionality to examine results for different water quality parameters in Pensacola and Perdido Bays
Install and import the required libraries
[1]:
import sys
#!python -m pip uninstall harmonize-wq --yes
#!python -m pip install harmonize-wq --yes
# Use pip to install the package from pypi or the latest from github
#!{sys.executable} -m pip install harmonize-wq
# For latest dev version
#!{sys.executable} -m pip install git+https://github.com/USEPA/harmonize-wq.git@new_release_0-3-8
[2]:
import dataretrieval.wqp as wqp
from harmonize_wq import wrangle
from harmonize_wq import location
from harmonize_wq import harmonize
from harmonize_wq import visualize
from harmonize_wq import clean
Download location data using dataretrieval
[3]:
# Read geometry for Area of Interest from geojson file url and plot
aoi_url = r'https://raw.githubusercontent.com/USEPA/harmonize-wq/main/harmonize_wq/tests/data/PPBays_NCCA.geojson'
aoi_gdf = wrangle.as_gdf(aoi_url).to_crs(epsg=4326) # already standard 4326
aoi_gdf.plot()
[3]:
<Axes: >
[4]:
# Note there are actually two polygons (one for each Bay)
aoi_gdf
# Spatial query parameters can be updated to run just one
bBox = wrangle.get_bounding_box(aoi_gdf)
# For only one bay, e.g., first is Pensacola Bay:
#bBox = wrangle.get_bounding_box(aoi_gdf, 0)
[5]:
# Build query with characteristicNames and the AOI extent
query = {'characteristicName': ['Phosphorus',
'Temperature, water',
'Depth, Secchi disk depth',
'Dissolved oxygen (DO)',
'Salinity',
'pH',
'Nitrogen',
'Conductivity',
'Organic carbon',
'Chlorophyll a',
'Turbidity',
'Sediment',
'Fecal Coliform',
'Escherichia coli']}
query['bBox'] = bBox
[6]:
# Query stations (can be slow)
stations, site_md = wqp.what_sites(**query)
[7]:
# Rows and columns for results
stations.shape
[7]:
(2938, 37)
[8]:
# First 5 rows
stations.head()
[8]:
| OrganizationIdentifier | OrganizationFormalName | MonitoringLocationIdentifier | MonitoringLocationName | MonitoringLocationTypeName | MonitoringLocationDescriptionText | HUCEightDigitCode | DrainageAreaMeasure/MeasureValue | DrainageAreaMeasure/MeasureUnitCode | ContributingDrainageAreaMeasure/MeasureValue | ... | AquiferName | LocalAqfrName | FormationTypeText | AquiferTypeName | ConstructionDateText | WellDepthMeasure/MeasureValue | WellDepthMeasure/MeasureUnitCode | WellHoleDepthMeasure/MeasureValue | WellHoleDepthMeasure/MeasureUnitCode | ProviderName | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | USGS-AL | USGS Alabama Water Science Center | USGS-02376115 | ELEVENMILE CREEK NR WEST PENSACOLA, FL | Stream | NaN | 3140107.0 | 27.8 | sq mi | 27.8 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NWIS |
| 1 | USGS-AL | USGS Alabama Water Science Center | USGS-02377570 | STYX RIVER NEAR ELSANOR, AL. | Stream | NaN | 3140106.0 | 192.0 | sq mi | 192.0 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NWIS |
| 2 | USGS-AL | USGS Alabama Water Science Center | USGS-02377920 | BLACKWATER RIVER AT US HWY 90 NR ROBERTSDALE, AL. | Stream | NaN | 3140106.0 | 23.1 | sq mi | 23.1 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NWIS |
| 3 | USGS-AL | USGS Alabama Water Science Center | USGS-02377960 | BLACKWATER RIVER AT CO RD 87 NEAR ELSANOR, AL. | Stream | NaN | 3140106.0 | 56.6 | sq mi | 56.6 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NWIS |
| 4 | USGS-AL | USGS Alabama Water Science Center | USGS-02377975 | BLACKWATER RIVER ABOVE SEMINOLE AL | Stream | NaN | 3140106.0 | 40.2 | sq mi | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NWIS |
5 rows × 37 columns
[9]:
# Columns used for an example row
stations.iloc[0][['HorizontalCoordinateReferenceSystemDatumName', 'LatitudeMeasure', 'LongitudeMeasure']]
[9]:
HorizontalCoordinateReferenceSystemDatumName NAD83
LatitudeMeasure 30.498252
LongitudeMeasure -87.335809
Name: 0, dtype: object
[10]:
# Harmonize location datums to 4326 (Note we keep intermediate columns using intermediate_columns=True)
stations_gdf = location.harmonize_locations(stations, out_EPSG=4326, intermediate_columns=True)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/clean.py:356: FutureWarning: Logical ops (and, or, xor) between Pandas objects and dtype-less sequences (e.g. list, tuple) are deprecated and will raise in a future version. Wrap the object in a Series, Index, or np.array before operating instead.
cond_notna = mask & (df_out["QA_flag"].notna()) # Mask cond and not NA
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/clean.py:360: FutureWarning: Logical ops (and, or, xor) between Pandas objects and dtype-less sequences (e.g. list, tuple) are deprecated and will raise in a future version. Wrap the object in a Series, Index, or np.array before operating instead.
df_out.loc[mask & (df_out["QA_flag"].isna()), "QA_flag"] = flag
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/clean.py:360: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value 'LatitudeMeasure: Imprecise: lessthan3decimaldigits' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[mask & (df_out["QA_flag"].isna()), "QA_flag"] = flag
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/clean.py:356: FutureWarning: Logical ops (and, or, xor) between Pandas objects and dtype-less sequences (e.g. list, tuple) are deprecated and will raise in a future version. Wrap the object in a Series, Index, or np.array before operating instead.
cond_notna = mask & (df_out["QA_flag"].notna()) # Mask cond and not NA
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/clean.py:360: FutureWarning: Logical ops (and, or, xor) between Pandas objects and dtype-less sequences (e.g. list, tuple) are deprecated and will raise in a future version. Wrap the object in a Series, Index, or np.array before operating instead.
df_out.loc[mask & (df_out["QA_flag"].isna()), "QA_flag"] = flag
[11]:
location.harmonize_locations?
[12]:
# Rows and columns for results after running the function (5 new columns, only 2 new if intermediate_columns=False)
stations_gdf.shape
[12]:
(2938, 42)
[13]:
# Example results for the new columns
stations_gdf.iloc[0][['geom_orig', 'EPSG', 'QA_flag', 'geom', 'geometry']]
[13]:
geom_orig (-87.3358086, 30.49825159)
EPSG 4269.0
QA_flag NaN
geom POINT (-87.3358086 30.49825159)
geometry POINT (-87.3358086 30.49825159)
Name: 0, dtype: object
[14]:
# geom and geometry look the same but geometry is a special datatype
stations_gdf['geometry'].dtype
[14]:
<geopandas.array.GeometryDtype at 0x7fd97891dfa0>
[15]:
# Look at the different QA_flag flags that have been assigned,
# e.g., for bad datums or limited decimal precision
set(stations_gdf.loc[stations_gdf['QA_flag'].notna()]['QA_flag'])
[15]:
{'HorizontalCoordinateReferenceSystemDatumName: Bad datum OTHER, EPSG:4326 assumed',
'HorizontalCoordinateReferenceSystemDatumName: Bad datum UNKWN, EPSG:4326 assumed',
'LatitudeMeasure: Imprecise: lessthan3decimaldigits',
'LatitudeMeasure: Imprecise: lessthan3decimaldigits; HorizontalCoordinateReferenceSystemDatumName: Bad datum UNKWN, EPSG:4326 assumed',
'LatitudeMeasure: Imprecise: lessthan3decimaldigits; LongitudeMeasure: Imprecise: lessthan3decimaldigits',
'LongitudeMeasure: Imprecise: lessthan3decimaldigits',
'LongitudeMeasure: Imprecise: lessthan3decimaldigits; HorizontalCoordinateReferenceSystemDatumName: Bad datum UNKWN, EPSG:4326 assumed'}
[16]:
# Map it
stations_gdf.plot()
[16]:
<Axes: >
[17]:
# Clip to area of interest
stations_clipped = wrangle.clip_stations(stations_gdf, aoi_gdf)
[18]:
# Map it
stations_clipped.plot()
[18]:
<Axes: >
[19]:
# How many stations now?
len(stations_clipped)
[19]:
1476
[20]:
# To save the results to a shapefile
#import os
#path = '' #specify the path (folder/directory) to save it to
#stations_clipped.to_file(os.path.join(path, 'PPBEP_stations.shp'))
Retrieve Characteristic Data
[21]:
# Now query for results
query['dataProfile'] = 'narrowResult'
res_narrow, md_narrow = wqp.get_results(**query)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/dataretrieval/wqp.py:153: DtypeWarning: Columns (10,13,15,17,19,20,21,22,23,28,31,33,34,36,58,60,61,64,65,69,70,71,72,73) have mixed types. Specify dtype option on import or set low_memory=False.
df = pd.read_csv(StringIO(response.text), delimiter=",")
[22]:
df = res_narrow
df
[22]:
| OrganizationIdentifier | OrganizationFormalName | ActivityIdentifier | ActivityStartDate | ActivityStartTime/Time | ActivityStartTime/TimeZoneCode | MonitoringLocationIdentifier | ResultIdentifier | DataLoggerLine | ResultDetectionConditionText | ... | AnalysisEndTime/TimeZoneCode | ResultLaboratoryCommentCode | ResultLaboratoryCommentText | ResultDetectionQuantitationLimitUrl | LaboratoryAccreditationIndicator | LaboratoryAccreditationAuthorityName | TaxonomistAccreditationIndicator | TaxonomistAccreditationAuthorityName | LabSamplePreparationUrl | ProviderName | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | AWW_WQX | Alabama Water Watch | AWW_WQX-aww_0321:20131111121500:SR:WSO | 2013-11-11 | 12:15:00 | CST | AWW_WQX-aww_0321 | STORET-1079479903 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | STORET |
| 1 | 21FLSEAS_WQX | Florida Department of Environmental Protection | 21FLSEAS_WQX-027950424132 | 2013-04-24 | 09:01:00 | EST | 21FLSEAS_WQX-02SEAS795 | STORET-310551339 | NaN | NaN | ... | NaN | NaN | NaN | https://www.waterqualitydata.us/data/providers... | NaN | NaN | NaN | NaN | NaN | STORET |
| 2 | 21FLSEAS_WQX | Florida Department of Environmental Protection | 21FLSEAS_WQX-027400613134 | 2013-06-13 | 10:01:00 | EST | 21FLSEAS_WQX-02SEAS740 | STORET-310489836 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | STORET |
| 3 | 21FLPNS_WQX | FL Dept. of Environmental Protection, Northwes... | 21FLPNS_WQX-1536988F1 | 2013-09-17 | 11:01:00 | EST | 21FLPNS_WQX-33030019 | STORET-308146602 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | STORET |
| 4 | AWW_WQX | Alabama Water Watch | AWW_WQX-aww_0330:20130112134500:SR:WSO | 2013-01-12 | 13:45:00 | CST | AWW_WQX-aww_0330 | STORET-1079461086 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | STORET |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 463839 | USGS-AL | USGS Alabama Water Science Center | nwisal.01.99900500 | 1999-03-02 | 14:20:00 | CST | USGS-02376115 | NWIS-104002666 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NWIS |
| 463840 | USGS-AL | USGS Alabama Water Science Center | nwisal.01.00201479 | 2001-11-28 | 12:05:00 | CST | USGS-02377570 | NWIS-53918846 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NWIS |
| 463841 | USGS-AL | USGS Alabama Water Science Center | nwisal.01.00202076 | 2001-10-03 | 16:40:00 | CDT | USGS-02376115 | NWIS-104000948 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NWIS |
| 463842 | USGS-AL | USGS Alabama Water Science Center | nwisal.01.00202072 | 2001-11-28 | 13:45:00 | CST | USGS-02376115 | NWIS-104000936 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NWIS |
| 463843 | USGS-AL | USGS Alabama Water Science Center | nwisal.01.00201474 | 2001-10-03 | 14:15:00 | CDT | USGS-02377570 | NWIS-53918826 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NWIS |
463844 rows × 78 columns
[23]:
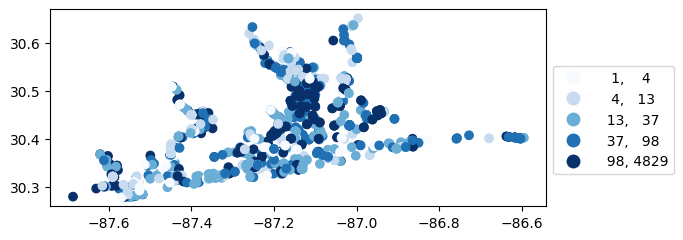
# Map number of usable results at each station
gdf_count = visualize.map_counts(df, stations_clipped)
legend_kwds = {"fmt": "{:.0f}", 'bbox_to_anchor':(1, 0.75)}
gdf_count.plot(column='cnt', cmap='Blues', legend=True, scheme='quantiles', legend_kwds=legend_kwds)
[23]:
<Axes: >
Harmonize Characteristic Results
Two options for functions to harmonize characteristics: harmonize_all() or harmonize_generic(). harmonize_all runs functions on all characteristics and lets you specify how to handle errors harmonize_generic runs functions only on the characteristic specified with char_val and lets you also choose output units, to keep intermediate columns and to do a quick report summarizing changes.
[24]:
# See Documentation
#harmonize.harmonize_all?
#harmonize.harmonize?
secchi disk depth
[25]:
# Each harmonize function has optional params, e.g., char_val is the characticName column value to use so we can send the entire df.
# Optional params: units='m', char_val='Depth, Secchi disk depth', out_col='Secchi', report=False)
# We start by demonstrating on secchi disk depth (units default to m, keep intermediate fields, see report)
df = harmonize.harmonize(df, 'Depth, Secchi disk depth', intermediate_columns=True, report=True)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/clean.py:360: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value 'ResultMeasureValue: "Not Reported" result cannot be used' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[mask & (df_out["QA_flag"].isna()), "QA_flag"] = flag
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(2.0, 'meter')> <Quantity(0.94, 'meter')>
<Quantity(0.6, 'meter')> ... <Quantity(1.67, 'meter')>
<Quantity(0.3048, 'meter')> <Quantity(0.11, 'meter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
-Usable results-
count 19538.000000
mean 1.166685
std 2.026694
min 0.000000
25% 0.600000
50% 1.000000
75% 1.400000
max 260.000000
dtype: float64
Unusable results: 88
Usable results with inferred units: 0
Results outside threshold (0.0 to 13.326848441851071): 1
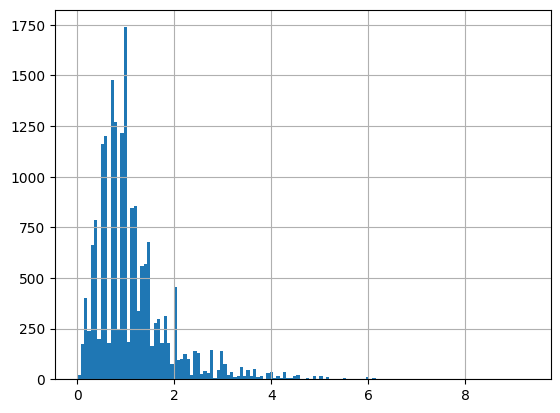
The threshold is based on standard deviations and is currently only used in the histogram.
[26]:
# Look at a table of just Secchi results and focus on subset of columns
cols = ['MonitoringLocationIdentifier', 'ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Units']
sechi_results = df.loc[df['CharacteristicName']=='Depth, Secchi disk depth', cols + ['Secchi']]
sechi_results
[26]:
| MonitoringLocationIdentifier | ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Units | Secchi | |
|---|---|---|---|---|---|---|
| 4 | AWW_WQX-aww_0330 | 2 | m | NaN | m | 2.0 meter |
| 99 | 21AWIC-7290 | .94 | m | NaN | m | 0.94 meter |
| 111 | 21FLPNS_WQX-33020JF1 | 0.60 | m | NaN | m | 0.6 meter |
| 131 | 21FLGW_WQX-3565 | .3 | m | NaN | m | 0.3 meter |
| 143 | 21FLBFA_WQX-33010016 | 1.5 | m | NaN | m | 1.5 meter |
| ... | ... | ... | ... | ... | ... | ... |
| 462625 | 21FLPNS_WQX-G4NW0441 | 0.8 | m | NaN | m | 0.8 meter |
| 462653 | 21FLPNS_WQX-3302H32GS1 | 0.7925 | m | NaN | m | 0.7925 meter |
| 462672 | 21FLESC_WQX-548AC-24Q4A | 1.67 | m | NaN | m | 1.67 meter |
| 462675 | 21FLPNS_WQX-3302J1GS7 | 0.3048 | m | NaN | m | 0.3048 meter |
| 462676 | 21FLGW_WQX-3565 | 0.11 | m | NaN | m | 0.11 meter |
19626 rows × 6 columns
[27]:
# Look at unusable(NAN) results
sechi_results.loc[df['Secchi'].isna()]
[27]:
| MonitoringLocationIdentifier | ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Units | Secchi | |
|---|---|---|---|---|---|---|
| 125369 | 21FLKWAT_WQX-OKA-CBA-GAP-3-1 | Not Reported | NaN | ResultMeasureValue: "Not Reported" result cann... | m | NaN |
| 125383 | 21FLCBA_WQX-OKA-CB-BASS-1 | Not Reported | NaN | ResultMeasureValue: "Not Reported" result cann... | m | NaN |
| 125732 | 21FLCBA_WQX-OKA-CBA-GAP-3-1 | Not Reported | NaN | ResultMeasureValue: "Not Reported" result cann... | m | NaN |
| 125949 | 21FLCBA_WQX-OKA-CB-BASS-1 | Not Reported | NaN | ResultMeasureValue: "Not Reported" result cann... | m | NaN |
| 127122 | 21FLKWAT_WQX-OKA-CB-BASS-1 | Not Reported | NaN | ResultMeasureValue: "Not Reported" result cann... | m | NaN |
| ... | ... | ... | ... | ... | ... | ... |
| 318266 | 21FLKWAT_WQX-OKA-CB-BASS-2 | Not Reported | ft | ResultMeasureValue: "Not Reported" result cann... | ft | NaN |
| 320506 | 21FLCBA_WQX-OKA-CB-BASS-1 | Not Reported | NaN | ResultMeasureValue: "Not Reported" result cann... | m | NaN |
| 325588 | 21FLCBA_WQX-OKA-CB-BASS-2 | Not Reported | NaN | ResultMeasureValue: "Not Reported" result cann... | m | NaN |
| 368897 | 21FLPNS_WQX-3302M13G | *Not Reported | m | ResultMeasureValue: "*Not Reported" result can... | m | NaN |
| 462197 | 21FLKWAT_WQX-OKA-CB-BASS-2 | Not Reported | ft | ResultMeasureValue: "Not Reported" result cann... | ft | NaN |
88 rows × 6 columns
[28]:
# look at the QA flag for first row from above
list(sechi_results.loc[df['Secchi'].isna()]['QA_flag'])[0]
[28]:
'ResultMeasureValue: "Not Reported" result cannot be used; ResultMeasure/MeasureUnitCode: MISSING UNITS, m assumed'
[29]:
# All cases where there was a QA flag
sechi_results.loc[df['QA_flag'].notna()]
[29]:
| MonitoringLocationIdentifier | ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Units | Secchi | |
|---|---|---|---|---|---|---|
| 125369 | 21FLKWAT_WQX-OKA-CBA-GAP-3-1 | Not Reported | NaN | ResultMeasureValue: "Not Reported" result cann... | m | NaN |
| 125383 | 21FLCBA_WQX-OKA-CB-BASS-1 | Not Reported | NaN | ResultMeasureValue: "Not Reported" result cann... | m | NaN |
| 125732 | 21FLCBA_WQX-OKA-CBA-GAP-3-1 | Not Reported | NaN | ResultMeasureValue: "Not Reported" result cann... | m | NaN |
| 125949 | 21FLCBA_WQX-OKA-CB-BASS-1 | Not Reported | NaN | ResultMeasureValue: "Not Reported" result cann... | m | NaN |
| 127122 | 21FLKWAT_WQX-OKA-CB-BASS-1 | Not Reported | NaN | ResultMeasureValue: "Not Reported" result cann... | m | NaN |
| ... | ... | ... | ... | ... | ... | ... |
| 318266 | 21FLKWAT_WQX-OKA-CB-BASS-2 | Not Reported | ft | ResultMeasureValue: "Not Reported" result cann... | ft | NaN |
| 320506 | 21FLCBA_WQX-OKA-CB-BASS-1 | Not Reported | NaN | ResultMeasureValue: "Not Reported" result cann... | m | NaN |
| 325588 | 21FLCBA_WQX-OKA-CB-BASS-2 | Not Reported | NaN | ResultMeasureValue: "Not Reported" result cann... | m | NaN |
| 368897 | 21FLPNS_WQX-3302M13G | *Not Reported | m | ResultMeasureValue: "*Not Reported" result can... | m | NaN |
| 462197 | 21FLKWAT_WQX-OKA-CB-BASS-2 | Not Reported | ft | ResultMeasureValue: "Not Reported" result cann... | ft | NaN |
88 rows × 6 columns
If both value and unit are missing nothing can be done, a unitless (NaN) value is assumed as to be in default units but a QA_flag is added
[30]:
# Aggregate Secchi data by station
visualize.station_summary(sechi_results, 'Secchi')
[30]:
| MonitoringLocationIdentifier | cnt | mean | |
|---|---|---|---|
| 0 | 11NPSWRD_WQX-GUIS_CMP_PKT01 | 12 | 2.333333 |
| 1 | 11NPSWRD_WQX-GUIS_CMP_PKT02 | 17 | 2.411765 |
| 2 | 11NPSWRD_WQX-GUIS_CMP_PKT03 | 3 | 2.333333 |
| 3 | 21AWIC-1063 | 124 | 0.775726 |
| 4 | 21AWIC-1122 | 64 | 2.981156 |
| ... | ... | ... | ... |
| 1168 | NARS_WQX-NCCA10-1432 | 1 | 1.075000 |
| 1169 | NARS_WQX-NCCA10-1433 | 1 | 1.423333 |
| 1170 | NARS_WQX-NCCA10-1434 | 1 | 2.400000 |
| 1171 | NARS_WQX-NCCA10-1488 | 1 | 0.736667 |
| 1172 | NARS_WQX-NCCA10-2432 | 1 | 1.600000 |
1173 rows × 3 columns
[31]:
# Map number of usable results at each station
gdf_count = visualize.map_counts(sechi_results, stations_clipped)
gdf_count.plot(column='cnt', cmap='Blues', legend=True, scheme='quantiles', legend_kwds=legend_kwds)
[31]:
<Axes: >
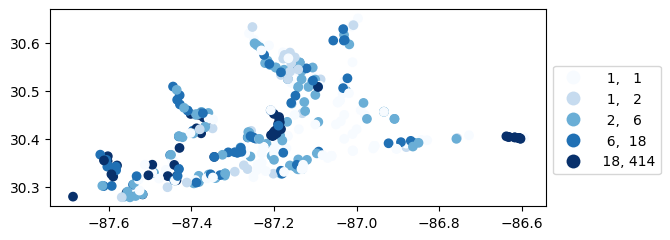
[32]:
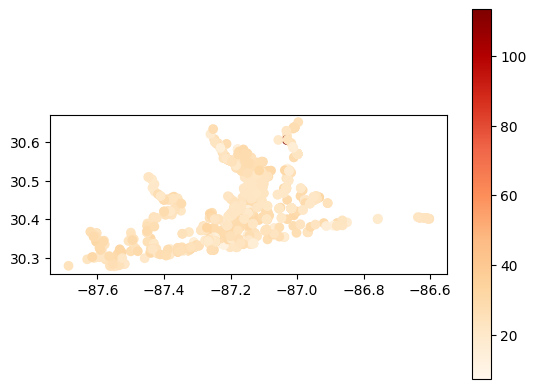
# Map average secchi depth results at each station
gdf_avg = visualize.map_measure(sechi_results, stations_clipped, 'Secchi')
gdf_avg.plot(column='mean', cmap='OrRd', legend=True)
[32]:
<Axes: >
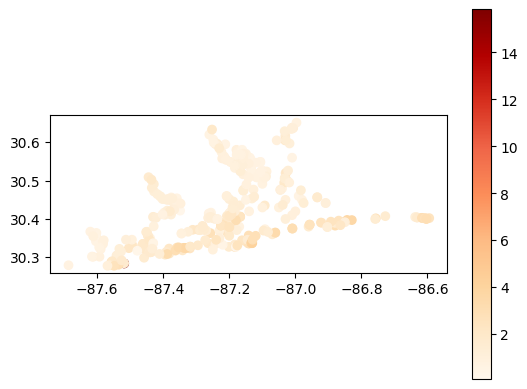
Temperature
The default error=’raise’, makes it so that there is an error when there is a dimensionality error (i.e. when units can’t be converted). Here we would get the error: DimensionalityError: Cannot convert from ‘count’ (dimensionless) to ‘degree_Celsius’ ([temperature])
[33]:
#'Temperature, water'
# errors=‘ignore’, invalid dimension conversions will return the NaN.
df = harmonize.harmonize(df, 'Temperature, water', intermediate_columns=True, report=True, errors='ignore')
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(26.0555556, 'degree_Celsius')>
<Quantity(12.35, 'degree_Celsius')> <Quantity(23.0, 'degree_Celsius')>
... <Quantity(25.0, 'degree_Celsius')> <Quantity(24.0, 'degree_Celsius')>
<Quantity(20.5, 'degree_Celsius')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
-Usable results-
count 94884.000000
mean 22.052907
std 9.858026
min -12.944444
25% 17.100000
50% 22.300000
75% 27.200000
max 1876.000000
dtype: float64
Unusable results: 2
Usable results with inferred units: 10
Results outside threshold (0.0 to 81.20106248802685): 10
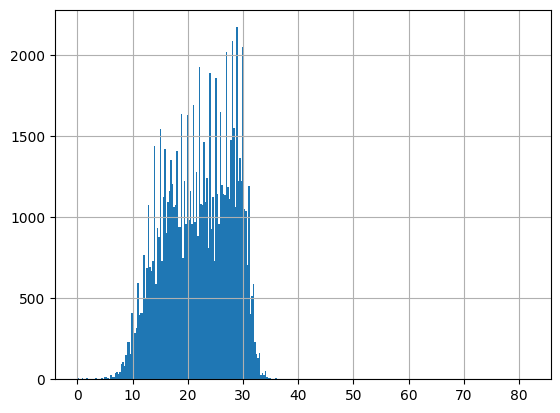
[34]:
# Look at what was changed
cols = ['MonitoringLocationIdentifier', 'ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Temperature', 'Units']
temperature_results = df.loc[df['CharacteristicName']=='Temperature, water', cols]
temperature_results
[34]:
| MonitoringLocationIdentifier | ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Temperature | Units | |
|---|---|---|---|---|---|---|
| 6 | 21FLCBA_WQX-BAS02 | 78.9 | deg F | NaN | 26.0555555555556 degree_Celsius | degF |
| 8 | 21FLPNS_WQX-33020J10 | 12.35 | deg C | NaN | 12.35 degree_Celsius | degC |
| 19 | AWW_WQX-aww_0318 | 23 | deg C | NaN | 23.0 degree_Celsius | degC |
| 21 | AWW_WQX-aww_1738 | 18.5 | deg C | NaN | 18.5 degree_Celsius | degC |
| 27 | 21FLSEAS_WQX-02SEAS810 | 23 | deg C | NaN | 23.0 degree_Celsius | degC |
| ... | ... | ... | ... | ... | ... | ... |
| 463839 | USGS-02376115 | 23.0 | deg C | NaN | 23.0 degree_Celsius | degC |
| 463840 | USGS-02377570 | 20.0 | deg C | NaN | 20.0 degree_Celsius | degC |
| 463841 | USGS-02376115 | 25.0 | deg C | NaN | 25.0 degree_Celsius | degC |
| 463842 | USGS-02376115 | 24.0 | deg C | NaN | 24.0 degree_Celsius | degC |
| 463843 | USGS-02377570 | 20.5 | deg C | NaN | 20.5 degree_Celsius | degC |
94886 rows × 6 columns
In the above we can see examples where the results were in deg F and in the result field they’ve been converted into degree_Celsius
[35]:
# Examine missing units
temperature_results.loc[df['ResultMeasure/MeasureUnitCode'].isna()]
[35]:
| MonitoringLocationIdentifier | ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Temperature | Units | |
|---|---|---|---|---|---|---|
| 188676 | NARS_WQX-OWW04440-0401 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN | degC |
| 255783 | 21FLCBA-RIV02 | 74.2 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 74.2 degree_Celsius | degC |
| 255788 | 21FLCBA-RIV02 | 74.2 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 74.2 degree_Celsius | degC |
| 256370 | 21FLCBA-FWB02 | 82.1 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 82.1 degree_Celsius | degC |
| 256371 | 21FLCBA-FWB02 | 82.6 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 82.6 degree_Celsius | degC |
| 256372 | 21FLCBA-FWB02 | 71.8 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 71.8 degree_Celsius | degC |
| 256373 | 21FLCBA-FWB02 | 79.4 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 79.4 degree_Celsius | degC |
| 257971 | 21FLCBA-FWB01 | 83.3 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 83.3 degree_Celsius | degC |
| 258796 | 21FLCBA-FWB05 | 79.8 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 79.8 degree_Celsius | degC |
| 259895 | 21FLCBA-FWB01 | 71.2 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 71.2 degree_Celsius | degC |
| 259900 | 21FLCBA-FWB05 | 81.7 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 81.7 degree_Celsius | degC |
We can see where the units were missing, the results were assumed to be in degree_Celsius already
[36]:
# This is also noted in the QA_flag field
list(temperature_results.loc[df['ResultMeasure/MeasureUnitCode'].isna(), 'QA_flag'])[0]
[36]:
'ResultMeasureValue: missing (NaN) result; ResultMeasure/MeasureUnitCode: MISSING UNITS, degC assumed'
[37]:
# Look for any without usable results
temperature_results.loc[df['Temperature'].isna()]
[37]:
| MonitoringLocationIdentifier | ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Temperature | Units | |
|---|---|---|---|---|---|---|
| 70456 | 11NPSWRD_WQX-GUIS_NALO | NaN | deg C | ResultMeasureValue: missing (NaN) result | NaN | degC |
| 188676 | NARS_WQX-OWW04440-0401 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN | degC |
[38]:
# Aggregate temperature data by station
visualize.station_summary(temperature_results, 'Temperature')
[38]:
| MonitoringLocationIdentifier | cnt | mean | |
|---|---|---|---|
| 0 | 11NPSWRD_WQX-GUIS_ADEM_ALPT | 30 | 24.986667 |
| 1 | 11NPSWRD_WQX-GUIS_BCCA | 1 | 36.800000 |
| 2 | 11NPSWRD_WQX-GUIS_BISA | 32 | 22.696250 |
| 3 | 11NPSWRD_WQX-GUIS_BOPI | 1 | 32.000000 |
| 4 | 11NPSWRD_WQX-GUIS_CMP_PKT01 | 20 | 25.125000 |
| ... | ... | ... | ... |
| 2544 | UWFCEDB_WQX-SRC-AI31-22 | 19 | 21.900000 |
| 2545 | UWFCEDB_WQX-SRC-AI36-22 | 26 | 21.957692 |
| 2546 | UWFCEDB_WQX-SRC-AI42-22 | 21 | 22.590476 |
| 2547 | UWFCEDB_WQX-SRC-AI44-22 | 24 | 21.095833 |
| 2548 | UWFCEDB_WQX-SRC-AK41-22 | 20 | 22.015000 |
2549 rows × 3 columns
[39]:
# Map number of usable results at each station
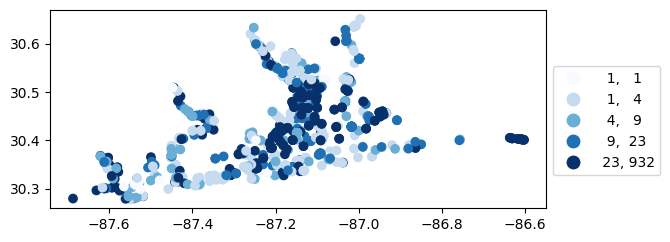
gdf_count = visualize.map_counts(temperature_results, stations_clipped)
gdf_count.plot(column='cnt', cmap='Blues', legend=True, scheme='quantiles', legend_kwds=legend_kwds)
[39]:
<Axes: >
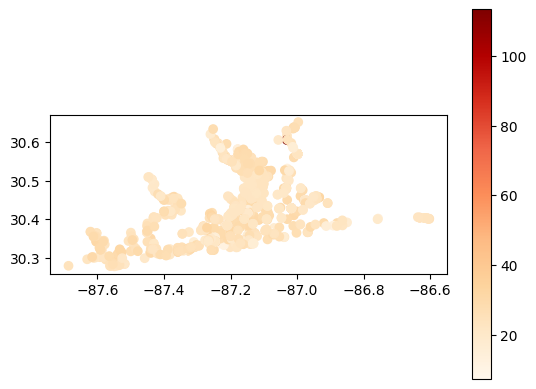
[40]:
# Map average temperature results at each station
gdf_temperature = visualize.map_measure(temperature_results, stations_clipped, 'Temperature')
gdf_temperature.plot(column='mean', cmap='OrRd', legend=True)
[40]:
<Axes: >
Dissolved oxygen
[41]:
# look at Dissolved oxygen (DO), but this time without intermediate fields
df = harmonize.harmonize(df, 'Dissolved oxygen (DO)')
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(6.3, 'milligram / liter')> <Quantity(4.5, 'milligram / liter')>
<Quantity(6.64, 'milligram / liter')> ...
<Quantity(6.6, 'milligram / liter')> <Quantity(7.0, 'milligram / liter')>
<Quantity(8.3, 'milligram / liter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
Note: Imediately when we run a harmonization function without the intermediate fields they’re deleted.
[42]:
# Look at what was changed
cols = ['MonitoringLocationIdentifier', 'ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'DO']
do_res = df.loc[df['CharacteristicName']=='Dissolved oxygen (DO)', cols]
do_res
[42]:
| MonitoringLocationIdentifier | ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | DO | |
|---|---|---|---|---|---|
| 2 | 21FLSEAS_WQX-02SEAS740 | 6.3 | mg/L | NaN | 6.3 milligram / liter |
| 7 | 21FLCMP_WQX-3201BM21 | 4.5 | mg/L | NaN | 4.5 milligram / liter |
| 15 | 21FLPNS_WQX-33030D71 | 6.64 | mg/L | NaN | 6.64 milligram / liter |
| 22 | 21FLBFA_WQX-33020057 | 1.17 | mg/L | NaN | 1.17 milligram / liter |
| 32 | 21FLNUTT_WQX-PB02 | 8.11 | mg/L | NaN | 8.11 milligram / liter |
| ... | ... | ... | ... | ... | ... |
| 463699 | 21AWIC-942 | 1.1 | mg/L | NaN | 1.1 milligram / liter |
| 463711 | 21AWIC-942 | 7.1 | mg/L | NaN | 7.1 milligram / liter |
| 463720 | 21AWIC-942 | 6.6 | mg/L | NaN | 6.6 milligram / liter |
| 463723 | 21AWIC-942 | 7 | mg/L | NaN | 7.0 milligram / liter |
| 463724 | 21AWIC-942 | 8.3 | mg/L | NaN | 8.3 milligram / liter |
74678 rows × 5 columns
[43]:
do_res.loc[do_res['ResultMeasure/MeasureUnitCode']!='mg/l']
[43]:
| MonitoringLocationIdentifier | ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | DO | |
|---|---|---|---|---|---|
| 2 | 21FLSEAS_WQX-02SEAS740 | 6.3 | mg/L | NaN | 6.3 milligram / liter |
| 7 | 21FLCMP_WQX-3201BM21 | 4.5 | mg/L | NaN | 4.5 milligram / liter |
| 15 | 21FLPNS_WQX-33030D71 | 6.64 | mg/L | NaN | 6.64 milligram / liter |
| 22 | 21FLBFA_WQX-33020057 | 1.17 | mg/L | NaN | 1.17 milligram / liter |
| 32 | 21FLNUTT_WQX-PB02 | 8.11 | mg/L | NaN | 8.11 milligram / liter |
| ... | ... | ... | ... | ... | ... |
| 463699 | 21AWIC-942 | 1.1 | mg/L | NaN | 1.1 milligram / liter |
| 463711 | 21AWIC-942 | 7.1 | mg/L | NaN | 7.1 milligram / liter |
| 463720 | 21AWIC-942 | 6.6 | mg/L | NaN | 6.6 milligram / liter |
| 463723 | 21AWIC-942 | 7 | mg/L | NaN | 7.0 milligram / liter |
| 463724 | 21AWIC-942 | 8.3 | mg/L | NaN | 8.3 milligram / liter |
51600 rows × 5 columns
Though there were no results in %, the conversion from percent saturation (%) to mg/l is special. This equation is being improved by integrating tempertaure and pressure instead of assuming STP (see DO_saturation())
[44]:
# Aggregate DO data by station
visualize.station_summary(do_res, 'DO')
[44]:
| MonitoringLocationIdentifier | cnt | mean | |
|---|---|---|---|
| 0 | 11NPSWRD_WQX-GUIS_ADEM_ALPT | 30 | 6.698000 |
| 1 | 11NPSWRD_WQX-GUIS_BCCA | 1 | 0.270000 |
| 2 | 11NPSWRD_WQX-GUIS_BISA | 32 | 7.194375 |
| 3 | 11NPSWRD_WQX-GUIS_BOPI | 1 | 7.540000 |
| 4 | 11NPSWRD_WQX-GUIS_FPPO | 1 | 9.950000 |
| ... | ... | ... | ... |
| 2154 | UWFCEDB_WQX-SRC-AI31-22 | 38 | 3.760918 |
| 2155 | UWFCEDB_WQX-SRC-AI36-22 | 52 | 3.514965 |
| 2156 | UWFCEDB_WQX-SRC-AI42-22 | 42 | 3.704803 |
| 2157 | UWFCEDB_WQX-SRC-AI44-22 | 48 | 3.798289 |
| 2158 | UWFCEDB_WQX-SRC-AK41-22 | 40 | 2.455314 |
2159 rows × 3 columns
[45]:
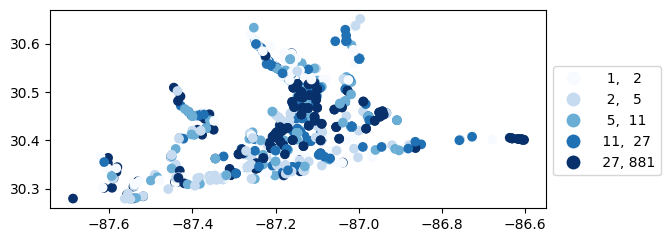
# Map number of usable results at each station
gdf_count = visualize.map_counts(do_res, stations_clipped)
gdf_count.plot(column='cnt', cmap='Blues', legend=True, scheme='quantiles', legend_kwds=legend_kwds)
[45]:
<Axes: >
[46]:
# Map Averages at each station
gdf_avg = visualize.map_measure(do_res, stations_clipped, 'DO')
gdf_avg.plot(column='mean', cmap='OrRd', legend=True)
[46]:
<Axes: >
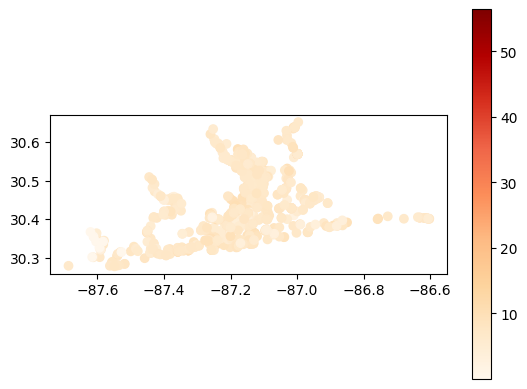
pH
[47]:
# pH, this time looking at a report
df = harmonize.harmonize(df, 'pH', report=True)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(7.29, 'dimensionless')> <Quantity(8.09, 'dimensionless')>
<Quantity(7.45, 'dimensionless')> ... <Quantity(8.47, 'dimensionless')>
<Quantity(8.48, 'dimensionless')> <Quantity(4.4, 'dimensionless')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
-Usable results-
count 59314.000000
mean 7.335669
std 0.904333
min 0.500000
25% 6.840000
50% 7.670000
75% 8.000000
max 16.200000
dtype: float64
Unusable results: 51
Usable results with inferred units: 58293
Results outside threshold (0.0 to 12.76166981004167): 1
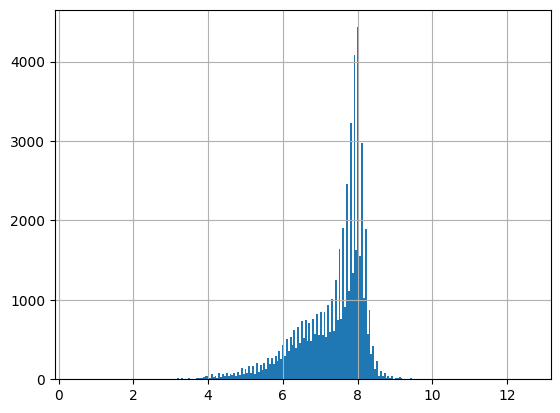
Note the warnings that occur when a unit is not recognized by the package. These occur even when report=False. Future versions could include these as defined units for pH, but here it wouldn’t alter results.
[48]:
df.loc[df['CharacteristicName']=='pH', ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'pH']]
[48]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | pH | |
|---|---|---|---|---|
| 3 | 7.29 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 7.29 dimensionless |
| 25 | 8.09 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 8.09 dimensionless |
| 30 | 7.45 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 7.45 dimensionless |
| 34 | 6.57 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 6.57 dimensionless |
| 36 | 6.57 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 6.57 dimensionless |
| ... | ... | ... | ... | ... |
| 463773 | 7 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 7.0 dimensionless |
| 463775 | 8.27 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 8.27 dimensionless |
| 463776 | 8.47 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 8.47 dimensionless |
| 463779 | 8.48 | NaN | ResultMeasure/MeasureUnitCode: MISSING UNITS, ... | 8.48 dimensionless |
| 463802 | 4.4 | std units | NaN | 4.4 dimensionless |
59365 rows × 4 columns
‘None’ is uninterpretable and replaced with NaN, which then gets replaced with ‘dimensionless’ since pH is unitless
Salinity
[49]:
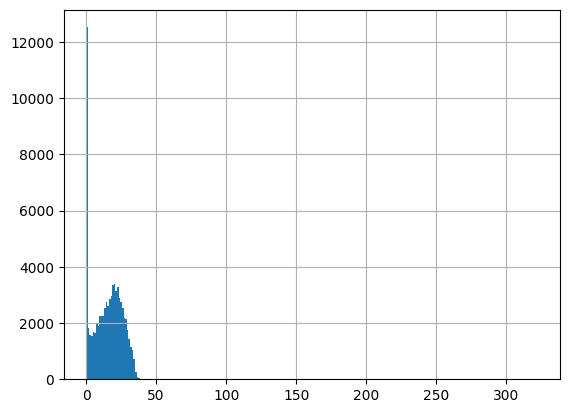
# Salinity
df = harmonize.harmonize(df, 'Salinity', report=True)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(1.012, 'Practical_Salinity_Units')>
<Quantity(18.9, 'Practical_Salinity_Units')>
<Quantity(25.0, 'Practical_Salinity_Units')> ...
<Quantity(2.11, 'Practical_Salinity_Units')>
<Quantity(1.89, 'Practical_Salinity_Units')>
<Quantity(2.12, 'Practical_Salinity_Units')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
-Usable results-
count 78445.000000
mean 15.733322
std 145.877352
min 0.000000
25% 5.790000
50% 15.990000
75% 23.100000
max 37782.000000
dtype: float64
Unusable results: 417
Usable results with inferred units: 10
Results outside threshold (0.0 to 890.9974365508828): 4
[50]:
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Salinity']
df.loc[df['CharacteristicName']=='Salinity', cols]
[50]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Salinity | |
|---|---|---|---|---|
| 0 | 1.012 | ppt | NaN | 1.012 Practical_Salinity_Units |
| 11 | 18.9 | ppth | NaN | 18.9 Practical_Salinity_Units |
| 12 | 25 | ppt | NaN | 25.0 Practical_Salinity_Units |
| 14 | 11.82 | ppth | NaN | 11.82 Practical_Salinity_Units |
| 23 | .03 | ppt | NaN | 0.03 Practical_Salinity_Units |
| ... | ... | ... | ... | ... |
| 463768 | 2.16 | ppth | NaN | 2.16 Practical_Salinity_Units |
| 463770 | 2.07 | ppth | NaN | 2.07 Practical_Salinity_Units |
| 463771 | 2.11 | ppth | NaN | 2.11 Practical_Salinity_Units |
| 463774 | 1.89 | ppth | NaN | 1.89 Practical_Salinity_Units |
| 463778 | 2.12 | ppth | NaN | 2.12 Practical_Salinity_Units |
78862 rows × 4 columns
Nitrogen
[51]:
# Nitrogen
df = harmonize.harmonize(df, 'Nitrogen', report=True)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/basis.py:343: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value 'as N' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[mask, basis_col] = basis
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:484: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '['as N' 'as N' 'as N' 'as N' 'as N' 'as N' 'as N' 'as N' 'as N' 'as N'
'as N' 'as N' 'as N' 'as N' 'as N' 'as N' 'as N' 'as N' 'as N' 'as N'
'as N' 'as N' nan 'as N' 'as N' nan nan nan nan 'as N' nan nan 'as N' nan
'as N' nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan
nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan
nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan
nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan
nan nan nan nan nan nan nan nan]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
self.df[c_mask] = basis.basis_from_method_spec(self.df[c_mask])
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:395: UserWarning: WARNING: 'cm3/g' UNDEFINED UNIT for Nitrogen
warn("WARNING: " + problem)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(1.5, 'milligram / liter')>
<Quantity(0.44, 'milligram / liter')>
<Quantity(0.68, 'milligram / liter')>
<Quantity(0.93, 'milligram / liter')>
<Quantity(0.26, 'milligram / liter')>
<Quantity(0.68, 'milligram / liter')>
<Quantity(0.26, 'milligram / liter')>
<Quantity(0.64, 'milligram / liter')>
<Quantity(1.1, 'milligram / liter')>
<Quantity(0.31, 'milligram / liter')>
<Quantity(1.0, 'milligram / liter')>
<Quantity(0.38, 'milligram / liter')>
<Quantity(1.7, 'milligram / liter')>
<Quantity(0.65, 'milligram / liter')>
<Quantity(0.3, 'milligram / liter')>
<Quantity(0.36, 'milligram / liter')>
<Quantity(0.33875, 'milligram / liter')>
<Quantity(0.53125, 'milligram / liter')>
<Quantity(135.0, 'milligram / liter')>
<Quantity(0.4075, 'milligram / liter')>
<Quantity(0.35375, 'milligram / liter')>
<Quantity(27.5, 'milligram / liter')>
<Quantity(82.4, 'milligram / liter')>
<Quantity(51.9, 'milligram / liter')>
<Quantity(11.8, 'milligram / liter')>
<Quantity(0.495, 'milligram / liter')>
<Quantity(131.0, 'milligram / liter')>
<Quantity(1630.0, 'milligram / liter')>
<Quantity(0.4475, 'milligram / liter')>
<Quantity(23.5, 'milligram / liter')>
<Quantity(0.36125, 'milligram / liter')>
<Quantity(49.8, 'milligram / liter')>
<Quantity(83.6, 'milligram / liter')>
<Quantity(197.0, 'milligram / liter')>
<Quantity(314.0, 'milligram / liter')>
<Quantity(0.636, 'milligram / liter')>
<Quantity(0.27, 'milligram / liter')>
<Quantity(0.86, 'milligram / liter')>
<Quantity(1.5, 'milligram / liter')>
<Quantity(0.87, 'milligram / liter')>
<Quantity(0.76, 'milligram / liter')>
<Quantity(1.12, 'milligram / liter')>
<Quantity(0.33, 'milligram / liter')>
<Quantity(1.3, 'milligram / liter')>
<Quantity(0.222, 'milligram / liter')>
<Quantity(0.37, 'milligram / liter')>
<Quantity(0.31724, 'milligram / liter')>
<Quantity(0.45668, 'milligram / liter')>
<Quantity(0.909, 'milligram / liter')>
<Quantity(0.67, 'milligram / liter')>
<Quantity(0.67, 'milligram / liter')>
<Quantity(1.13, 'milligram / liter')>
<Quantity(0.45906, 'milligram / liter')>
<Quantity(1.376, 'milligram / liter')>
<Quantity(0.3675, 'milligram / liter')>
<Quantity(1.2, 'milligram / liter')>
<Quantity(0.30226, 'milligram / liter')>
<Quantity(0.4263, 'milligram / liter')>
<Quantity(0.32, 'milligram / liter')>
<Quantity(0.531, 'milligram / liter')>
<Quantity(0.68, 'milligram / liter')>
<Quantity(0.61, 'milligram / liter')>
<Quantity(0.16, 'milligram / liter')>
<Quantity(0.55, 'milligram / liter')>
<Quantity(0.652, 'milligram / liter')>
<Quantity(0.629, 'milligram / liter')>
<Quantity(0.622, 'milligram / liter')>
<Quantity(0.62, 'milligram / liter')>
<Quantity(0.69, 'milligram / liter')>
<Quantity(0.62, 'milligram / liter')>
<Quantity(0.6, 'milligram / liter')>
<Quantity(0.57, 'milligram / liter')>
<Quantity(0.48986, 'milligram / liter')>
<Quantity(0.60326, 'milligram / liter')>
<Quantity(0.60368, 'milligram / liter')>
<Quantity(0.6, 'milligram / liter')>
<Quantity(0.77, 'milligram / liter')>
<Quantity(0.81, 'milligram / liter')>
<Quantity(0.57, 'milligram / liter')>
<Quantity(0.84, 'milligram / liter')>
<Quantity(0.86, 'milligram / liter')>
<Quantity(0.34846, 'milligram / liter')>
<Quantity(0.67, 'milligram / liter')>
<Quantity(0.96, 'milligram / liter')>
<Quantity(0.47642, 'milligram / liter')>
<Quantity(0.6, 'milligram / liter')>
<Quantity(0.48678, 'milligram / liter')>
<Quantity(0.5, 'milligram / liter')>
<Quantity(0.72, 'milligram / liter')>
<Quantity(0.41, 'milligram / liter')>
<Quantity(1.1, 'milligram / liter')>
<Quantity(0.65548, 'milligram / liter')>
<Quantity(0.3031, 'milligram / liter')>
<Quantity(0.52738, 'milligram / liter')>
<Quantity(0.27552, 'milligram / liter')>
<Quantity(0.28634, 'milligram / liter')>
<Quantity(0.5697, 'milligram / liter')>
<Quantity(0.0007, 'milligram / liter')>
<Quantity(0.0146, 'milligram / liter')>
<Quantity(0.0008, 'milligram / liter')>
<Quantity(0.0158, 'milligram / liter')>
<Quantity(16.46, 'milligram / liter')>
<Quantity(18.82, 'milligram / liter')>
<Quantity(17.76, 'milligram / liter')>
<Quantity(18.69, 'milligram / liter')>
<Quantity(16.18, 'milligram / liter')>
<Quantity(18.99, 'milligram / liter')>
<Quantity(18.72, 'milligram / liter')>
<Quantity(17.61, 'milligram / liter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/domains.py:277: FutureWarning: Downcasting object dtype arrays on .fillna, .ffill, .bfill is deprecated and will change in a future version. Call result.infer_objects(copy=False) instead. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
sub_df[cols[2]] = sub_df[cols[2]].fillna(sub_df[cols[1]]) # new_fract
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/domains.py:277: FutureWarning: Downcasting object dtype arrays on .fillna, .ffill, .bfill is deprecated and will change in a future version. Call result.infer_objects(copy=False) instead. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
sub_df[cols[2]] = sub_df[cols[2]].fillna(sub_df[cols[1]]) # new_fract
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/domains.py:277: FutureWarning: Downcasting object dtype arrays on .fillna, .ffill, .bfill is deprecated and will change in a future version. Call result.infer_objects(copy=False) instead. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
sub_df[cols[2]] = sub_df[cols[2]].fillna(sub_df[cols[1]]) # new_fract
-Usable results-
count 109.000000
mean 26.920174
std 160.257726
min 0.000700
25% 0.410000
50% 0.629000
75% 1.120000
max 1630.000000
dtype: float64
Unusable results: 4
Usable results with inferred units: 0
Results outside threshold (0.0 to 988.466532186079): 1
[52]:
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Nitrogen']
df.loc[df['CharacteristicName']=='Nitrogen', cols]
[52]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Nitrogen | |
|---|---|---|---|---|
| 57389 | 1.5 | mg/L | NaN | 1.5 milligram / liter |
| 58335 | 0.44 | mg/L | NaN | 0.44 milligram / liter |
| 59296 | 0.68 | mg/L | NaN | 0.68 milligram / liter |
| 59383 | 0.93 | mg/L | NaN | 0.93 milligram / liter |
| 60768 | 0.26 | mg/L | NaN | 0.26 milligram / liter |
| ... | ... | ... | ... | ... |
| 463262 | 18.69 | mg/l | NaN | 18.69 milligram / liter |
| 463269 | 16.18 | mg/l | NaN | 16.18 milligram / liter |
| 463270 | 18.99 | mg/l | NaN | 18.99 milligram / liter |
| 463273 | 18.72 | mg/l | NaN | 18.72 milligram / liter |
| 463275 | 17.61 | mg/l | NaN | 17.61 milligram / liter |
113 rows × 4 columns
Conductivity
[53]:
# Conductivity
df = harmonize.harmonize(df, 'Conductivity', report=True)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(19204.2, 'microsiemens / centimeter')>
<Quantity(222.3, 'microsiemens / centimeter')>
<Quantity(102.8, 'microsiemens / centimeter')> ...
<Quantity(78.0, 'microsiemens / centimeter')>
<Quantity(129.0, 'microsiemens / centimeter')>
<Quantity(72.0, 'microsiemens / centimeter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
-Usable results-
count 1818.000000
mean 17085.221414
std 16116.889030
min 0.040000
25% 130.000000
50% 16994.750000
75% 30306.650000
max 54886.200000
dtype: float64
Unusable results: 8
Usable results with inferred units: 0
Results outside threshold (0.0 to 113786.55559242623): 0
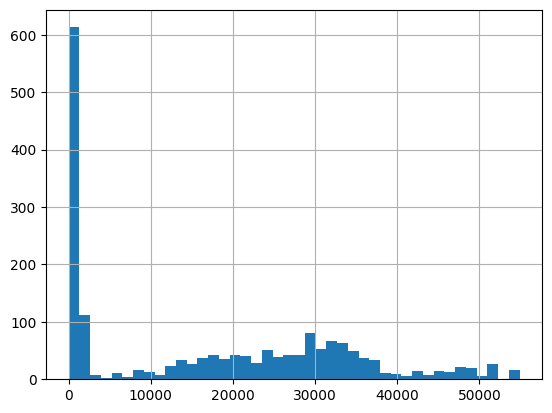
[54]:
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Conductivity']
df.loc[df['CharacteristicName']=='Conductivity', cols]
[54]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Conductivity | |
|---|---|---|---|---|
| 16 | 19204.2 | umho/cm | NaN | 19204.2 microsiemens / centimeter |
| 108 | 222.3 | umho/cm | NaN | 222.3 microsiemens / centimeter |
| 218 | 102.8 | umho/cm | NaN | 102.8 microsiemens / centimeter |
| 429 | 11017.5 | umho/cm | NaN | 11017.5 microsiemens / centimeter |
| 887 | 32 | umho/cm | NaN | 32.0 microsiemens / centimeter |
| ... | ... | ... | ... | ... |
| 463709 | 73 | umho/cm | NaN | 73.0 microsiemens / centimeter |
| 463712 | 77 | umho/cm | NaN | 77.0 microsiemens / centimeter |
| 463728 | 78 | umho/cm | NaN | 78.0 microsiemens / centimeter |
| 463734 | 129 | umho/cm | NaN | 129.0 microsiemens / centimeter |
| 463737 | 72 | umho/cm | NaN | 72.0 microsiemens / centimeter |
1826 rows × 4 columns
Chlorophyll a
[55]:
# Chlorophyll a
df = harmonize.harmonize(df, 'Chlorophyll a', report=True)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:395: UserWarning: WARNING: 'ug/cm2' UNDEFINED UNIT for Chlorophyll
warn("WARNING: " + problem)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(0.0023, 'milligram / liter')>
<Quantity(0.0029, 'milligram / liter')>
<Quantity(0.0041, 'milligram / liter')> ...
<Quantity(0.00672003521, 'milligram / liter')>
<Quantity(0.00229276774, 'milligram / liter')>
<Quantity(0.00500738688, 'milligram / liter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
-Usable results-
count 9464.000000
mean 1.145249
std 1.199159
min -0.840000
25% 0.007400
50% 0.940000
75% 1.820000
max 9.990000
dtype: float64
Unusable results: 631
Usable results with inferred units: 6175
Results outside threshold (0.0 to 8.34020536212113): 8
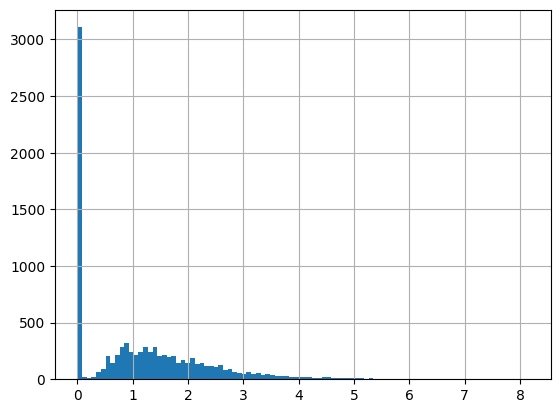
[56]:
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Chlorophyll']
df.loc[df['CharacteristicName']=='Chlorophyll a', cols]
[56]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Chlorophyll | |
|---|---|---|---|---|
| 277 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN |
| 618 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN |
| 736 | 2.3 | mg/m3 | NaN | 0.0023000000000000004 milligram / liter |
| 1351 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN |
| 1519 | 2.9 | mg/m3 | NaN | 0.0029000000000000007 milligram / liter |
| ... | ... | ... | ... | ... |
| 462433 | 9.43660497156775 | ug/L | NaN | 0.00943660497156775 milligram / liter |
| 462630 | 6.72003521191891 | ug/L | NaN | 0.0067200352119189104 milligram / liter |
| 462631 | 2.29276774101202 | ug/L | NaN | 0.00229276774101202 milligram / liter |
| 462640 | 5.00738687613263 | ug/L | NaN | 0.00500738687613263 milligram / liter |
| 463506 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN |
10095 rows × 4 columns
Organic Carbon
[57]:
# Organic carbon (%)
df = harmonize.harmonize(df, 'Organic carbon', report=True)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(5.4, 'milligram / liter')> <Quantity(2.6, 'milligram / liter')>
<Quantity(3.9, 'milligram / liter')> ...
<Quantity(1.7, 'milligram / liter')> <Quantity(1.5, 'milligram / liter')>
<Quantity(4.536, 'milligram / liter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
-Usable results-
count 5091.000000
mean 1079.369685
std 11287.026894
min 0.000000
25% 2.700000
50% 4.300000
75% 8.200000
max 410000.000000
dtype: float64
Unusable results: 165
Usable results with inferred units: 0
Results outside threshold (0.0 to 68801.53105006473): 22
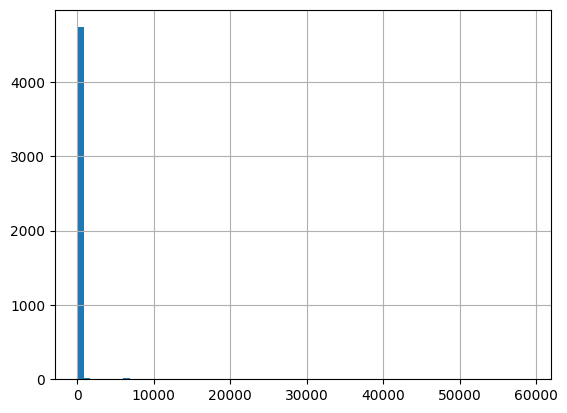
[58]:
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Carbon']
df.loc[df['CharacteristicName']=='Organic carbon', cols]
[58]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Carbon | |
|---|---|---|---|---|
| 140 | 5.4 | mg/L | NaN | 5.4 milligram / liter |
| 142 | 2.6 | mg/L | NaN | 2.6 milligram / liter |
| 178 | 3.9 | mg/L | NaN | 3.9 milligram / liter |
| 236 | 5.2 | mg/L | NaN | 5.2 milligram / liter |
| 296 | 6.0 | mg/L | NaN | 6.0 milligram / liter |
| ... | ... | ... | ... | ... |
| 463310 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN |
| 463311 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN |
| 463399 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN |
| 463407 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN |
| 463510 | 4.536 | mg/L | NaN | 4.536 milligram / liter |
5256 rows × 4 columns
Turbidity
[59]:
# Turbidity (NTU)
df = harmonize.harmonize(df, 'Turbidity', report=True)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(380.4523, 'Nephelometric_Turbidity_Units')>
<Quantity(0.0, 'Nephelometric_Turbidity_Units')>
<Quantity(190.2023, 'Nephelometric_Turbidity_Units')> ...
<Quantity(1.6, 'Nephelometric_Turbidity_Units')>
<Quantity(2.9, 'Nephelometric_Turbidity_Units')>
<Quantity(4.8, 'Nephelometric_Turbidity_Units')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
-Usable results-
count 47793.000000
mean 30.346572
std 205.790451
min -0.840000
25% 1.600000
50% 3.000000
75% 7.570000
max 32342.452300
dtype: float64
Unusable results: 610
Usable results with inferred units: 10
Results outside threshold (0.0 to 1265.0892804582636): 65
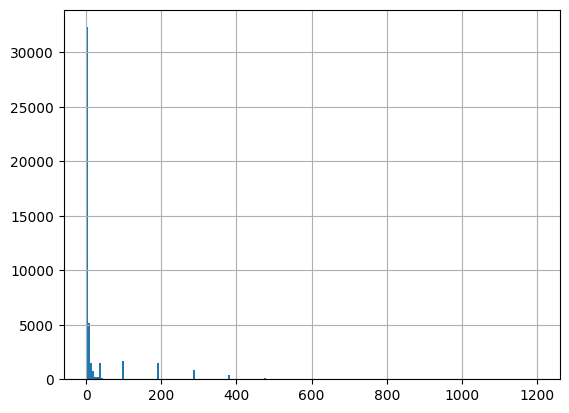
[60]:
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Turbidity']
df.loc[df['CharacteristicName']=='Turbidity', cols]
[60]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Turbidity | |
|---|---|---|---|---|
| 20 | 20 | JTU | NaN | 380.4523 Nephelometric_Turbidity_Units |
| 33 | 0 | NTU | NaN | 0.0 Nephelometric_Turbidity_Units |
| 46 | 10 | JTU | NaN | 190.2023 Nephelometric_Turbidity_Units |
| 56 | 1.4 | NTU | NaN | 1.4 Nephelometric_Turbidity_Units |
| 62 | 4.7 | NTU | NaN | 4.7 Nephelometric_Turbidity_Units |
| ... | ... | ... | ... | ... |
| 463701 | 3 | NTU | NaN | 3.0 Nephelometric_Turbidity_Units |
| 463715 | 4.7 | NTU | NaN | 4.7 Nephelometric_Turbidity_Units |
| 463716 | 1.6 | NTU | NaN | 1.6 Nephelometric_Turbidity_Units |
| 463718 | 2.9 | NTU | NaN | 2.9 Nephelometric_Turbidity_Units |
| 463727 | 4.8 | NTU | NaN | 4.8 Nephelometric_Turbidity_Units |
48403 rows × 4 columns
Sediment
[61]:
# Sediment
df = harmonize.harmonize(df, 'Sediment', report=False)
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
[62]:
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Sediment']
df.loc[df['CharacteristicName']=='Sediment', cols]
[62]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Sediment |
|---|
Phosphorus
Note: must be merged w/ activities (package runs query by site if not already merged)
[63]:
# Phosphorus
df = harmonize.harmonize(df, 'Phosphorus')
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[<Quantity(0.061, 'milligram / liter')>
<Quantity(0.03, 'milligram / liter')>
<Quantity(0.13, 'milligram / liter')> ...
<Quantity(0.16, 'milligram / liter')>
<Quantity(0.18, 'milligram / liter')>
<Quantity(0.18, 'milligram / liter')>]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
2 Phosphorus sample fractions not in frac_dict
2 Phosphorus sample fractions not in frac_dict found in expected domains, mapped to "Other_Phosphorus"
Note: warnings for unexpected characteristic fractions. Fractions are each seperated out into their own result column.
[64]:
# All Phosphorus
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'TDP_Phosphorus']
df.loc[df['Phosphorus'].notna(), cols]
[64]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | TDP_Phosphorus | |
|---|---|---|---|---|
| 45 | .061 | mg/L | NaN | NaN |
| 79 | 0.03 | mg/L | NaN | NaN |
| 174 | .13 | mg/L | NaN | NaN |
| 203 | 0.003 | mg/L | NaN | NaN |
| 355 | 0.002 | mg/L | NaN | NaN |
| ... | ... | ... | ... | ... |
| 463703 | .19 | mg/L | NaN | NaN |
| 463704 | .11 | mg/L | NaN | NaN |
| 463708 | .16 | mg/L | NaN | NaN |
| 463713 | .18 | mg/L | NaN | NaN |
| 463736 | .18 | mg/L | NaN | NaN |
7798 rows × 4 columns
[65]:
# Total phosphorus
df.loc[df['TP_Phosphorus'].notna(), cols]
[65]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | TDP_Phosphorus | |
|---|---|---|---|---|
| 45 | .061 | mg/L | NaN | NaN |
| 79 | 0.03 | mg/L | NaN | NaN |
| 174 | .13 | mg/L | NaN | NaN |
| 203 | 0.003 | mg/L | NaN | NaN |
| 355 | 0.002 | mg/L | NaN | NaN |
| ... | ... | ... | ... | ... |
| 463122 | 1.43 | mg/l as P | NaN | NaN |
| 463128 | 0.08 | mg/l as P | NaN | NaN |
| 463135 | 0.05 | mg/l as P | NaN | NaN |
| 463299 | 0.110 | mg/l as P | NaN | NaN |
| 463505 | .039 | mg/L | NaN | NaN |
6956 rows × 4 columns
[66]:
# Total dissolved phosphorus
df.loc[df['TDP_Phosphorus'].notna(), cols]
[66]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | TDP_Phosphorus | |
|---|---|---|---|---|
| 4208 | 0.019 | mg/L | NaN | 0.019 milligram / liter |
| 9433 | 0.002 | mg/L | NaN | 0.002 milligram / liter |
| 15788 | 0.003 | mg/L | NaN | 0.003 milligram / liter |
| 19109 | 0.019 | mg/L | NaN | 0.019 milligram / liter |
| 51079 | 0.002 | mg/L | NaN | 0.002 milligram / liter |
| 53187 | 0.017 | mg/L | NaN | 0.017 milligram / liter |
| 56084 | 0.020 | mg/L | NaN | 0.02 milligram / liter |
| 62492 | 0.002 | mg/L | NaN | 0.002 milligram / liter |
| 79601 | 0.021 | mg/L | NaN | 0.021 milligram / liter |
| 82681 | 0.003 | mg/L | NaN | 0.003 milligram / liter |
| 188233 | 0.00806 | mg/L | NaN | 0.00806 milligram / liter |
| 192252 | 0.000031 | mg/L | NaN | 3.1e-05 milligram / liter |
| 193690 | 0.002542 | mg/L | NaN | 0.002542 milligram / liter |
| 194389 | 0.00341 | mg/L | NaN | 0.00341 milligram / liter |
| 238332 | 0.00372 | mg/L | NaN | 0.00372 milligram / liter |
| 240768 | 0.00961 | mg/L | NaN | 0.00961 milligram / liter |
| 241871 | 0.00124 | mg/L | NaN | 0.00124 milligram / liter |
| 242885 | 0.01271 | mg/L | NaN | 0.01271 milligram / liter |
| 461312 | 0.030 | mg/l as P | NaN | 0.03 milligram / liter |
| 461321 | 0.033 | mg/l as P | NaN | 0.033 milligram / liter |
| 461324 | 0.024 | mg/l as P | NaN | 0.024 milligram / liter |
| 461330 | 0.028 | mg/l as P | NaN | 0.028 milligram / liter |
| 461339 | 0.021 | mg/l as P | NaN | 0.021 milligram / liter |
| 461344 | 0.023 | mg/l as P | NaN | 0.023 milligram / liter |
| 461356 | 0.037 | mg/l as P | NaN | 0.037 milligram / liter |
| 461417 | 0.023 | mg/l as P | NaN | 0.023 milligram / liter |
| 461425 | 0.02 | mg/l as P | NaN | 0.02 milligram / liter |
| 461441 | 0.04 | mg/l as P | NaN | 0.04 milligram / liter |
| 461456 | 0.03 | mg/l as P | NaN | 0.03 milligram / liter |
| 461465 | 0.025 | mg/l as P | NaN | 0.025 milligram / liter |
| 461492 | 0.05 | mg/l as P | NaN | 0.05 milligram / liter |
| 461501 | 0.15 | mg/l as P | NaN | 0.15 milligram / liter |
| 461521 | 0.03 | mg/l as P | NaN | 0.03 milligram / liter |
| 462792 | 0.02 | mg/l as P | NaN | 0.02 milligram / liter |
| 462814 | 0.07 | mg/l as P | NaN | 0.07 milligram / liter |
| 462822 | 0.08 | mg/l as P | NaN | 0.08 milligram / liter |
| 462837 | 0.02 | mg/l as P | NaN | 0.02 milligram / liter |
| 462857 | 0.02 | mg/l as P | NaN | 0.02 milligram / liter |
| 462869 | 0.04 | mg/l as P | NaN | 0.04 milligram / liter |
| 462889 | 0.02 | mg/l as P | NaN | 0.02 milligram / liter |
| 462902 | 0.05 | mg/l as P | NaN | 0.05 milligram / liter |
| 463129 | 0.03 | mg/l as P | NaN | 0.03 milligram / liter |
| 463136 | 0.05 | mg/l as P | NaN | 0.05 milligram / liter |
[67]:
# All other phosphorus sample fractions
df.loc[df['Other_Phosphorus'].notna(), cols]
[67]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | TDP_Phosphorus | |
|---|---|---|---|---|
| 26297 | .079 | mg/L | NaN | NaN |
| 26711 | .014 | mg/L | NaN | NaN |
| 27503 | .37 | mg/L | NaN | NaN |
| 28221 | .99 | mg/L | NaN | NaN |
| 28648 | .023 | mg/L | NaN | NaN |
| ... | ... | ... | ... | ... |
| 463703 | .19 | mg/L | NaN | NaN |
| 463704 | .11 | mg/L | NaN | NaN |
| 463708 | .16 | mg/L | NaN | NaN |
| 463713 | .18 | mg/L | NaN | NaN |
| 463736 | .18 | mg/L | NaN | NaN |
799 rows × 4 columns
Bacteria
Some equivalence assumptions are built-in where bacteria counts that are not equivalent are treated as such because there is no standard way to convert from one to another.
Fecal Coliform
[68]:
# Known unit with bad dimensionality ('Colony_Forming_Units * milliliter')
df = harmonize.harmonize(df, 'Fecal Coliform', report=True, errors='ignore')
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'MPN/100mL' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'cfu/100mL' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'CFU/100mL' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[nan nan nan ... nan nan nan]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
-Usable results-
count 10035.000000
mean 45.537618
std 448.839329
min 0.000000
25% 4.000000
50% 8.000000
75% 33.000000
max 33000.000000
dtype: float64
Unusable results: 40585
Usable results with inferred units: 0
Results outside threshold (0.0 to 2738.5735941387825): 6
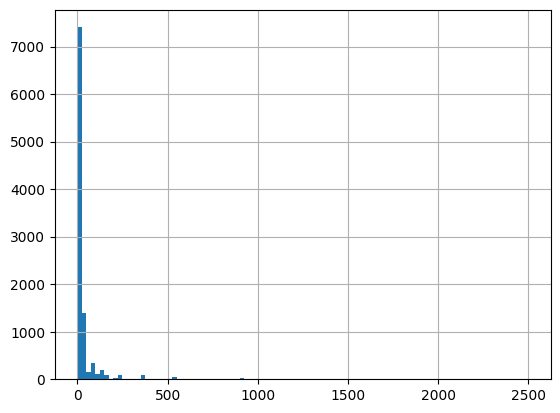
[69]:
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Fecal_Coliform']
df.loc[df['CharacteristicName']=='Fecal Coliform', cols]
[69]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Fecal_Coliform | |
|---|---|---|---|---|
| 1 | *Non-detect | NaN | ResultMeasureValue: "*Non-detect" result canno... | NaN |
| 5 | 80 | cfu/100mL | NaN | NaN |
| 9 | *Non-detect | NaN | ResultMeasureValue: "*Non-detect" result canno... | NaN |
| 10 | 2 | MPN/100mL | NaN | NaN |
| 13 | *Non-detect | NaN | ResultMeasureValue: "*Non-detect" result canno... | NaN |
| ... | ... | ... | ... | ... |
| 463681 | 300 | cfu/100mL | NaN | NaN |
| 463683 | 166 | cfu/100mL | NaN | NaN |
| 463719 | 190 | cfu/100mL | NaN | NaN |
| 463726 | 194 | cfu/100mL | NaN | NaN |
| 463732 | 226 | cfu/100mL | NaN | NaN |
50620 rows × 4 columns
Escherichia coli
[70]:
# Known unit with bad dimensionality ('Colony_Forming_Units * milliliter')
df = harmonize.harmonize(df, 'Escherichia coli', report=True, errors='ignore')
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:158: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
for bad_meas in pandas.unique(bad_measures):
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'MPN/100mL' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'cfu/100mL' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/convert.py:128: UserWarning: WARNING: 'CFU/100mL' converted to NaN
warn(f"WARNING: '{unit}' converted to NaN")
/opt/hostedtoolcache/Python/3.9.25/x64/lib/python3.9/site-packages/harmonize_wq/wq_data.py:663: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise an error in a future version of pandas. Value '[nan nan nan ... <Quantity(12.0, 'Colony_Forming_Units / milliliter')>
<Quantity(6.0, 'Colony_Forming_Units / milliliter')> nan]' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
df_out.loc[m_mask, self.out_col] = convert_unit_series(**params)
-Usable results-
count 22.000000
mean 501.863636
std 610.053260
min 4.000000
25% 9.500000
50% 77.500000
75% 1000.000000
max 1700.000000
dtype: float64
Unusable results: 11977
Usable results with inferred units: 0
Results outside threshold (0.0 to 4162.183198738116): 0
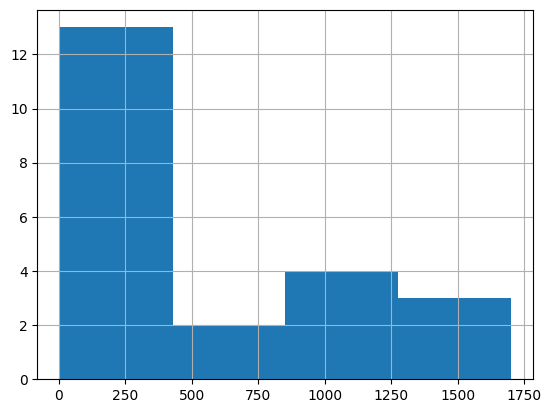
[71]:
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'E_coli']
df.loc[df['CharacteristicName']=='Escherichia coli', cols]
[71]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | E_coli | |
|---|---|---|---|---|
| 26 | 0 | cfu/100mL | NaN | NaN |
| 40 | 0 | cfu/100mL | NaN | NaN |
| 76 | 1000 | cfu/100mL | NaN | NaN |
| 82 | 33.3333333333333 | cfu/100mL | NaN | NaN |
| 87 | 0 | cfu/100mL | NaN | NaN |
| ... | ... | ... | ... | ... |
| 463280 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN |
| 463286 | NaN | NaN | ResultMeasureValue: missing (NaN) result; Resu... | NaN |
| 463401 | 12 | cfu/100ml | NaN | 12.0 Colony_Forming_Units / milliliter |
| 463409 | 6 | cfu/100ml | NaN | 6.0 Colony_Forming_Units / milliliter |
| 463513 | 1200 | MPN/100mL | NaN | NaN |
11999 rows × 4 columns
Combining Salinity and Conductivity
Convert module has various functions to convert from one unit or characteristic to another. Some of these are used within a single characteristic during harmonization (e.g. DO saturation to concentration) while others are intended to model one characteristic as an indicator of another (e.g. estimate salinity from conductivity).
Note: this should only be done after both characteristic fields have been harmonized. Results before and after should be inspected, thresholds for outliers applied, and consider adding a QA_flag for modeled data.
Explore Salinity results:
[72]:
from harmonize_wq import convert
[73]:
# Salinity summary statistics
lst = [x.magnitude for x in list(df['Salinity'].dropna())]
q_sum = sum(lst)
print('Range: {} to {}'.format(min(lst), max(lst)))
print('Results: {} \nMean: {} PSU'.format(len(lst), q_sum/len(lst)))
Range: 0.0 to 37782.0
Results: 78445
Mean: 15.733322044745321 PSU
[74]:
# Identify extreme outliers
[x for x in lst if x >3200]
[74]:
[15030.0, 37782.0]
Other fields like units and QA_flag may help understand what caused high values and what results might need to be dropped from consideration
[75]:
# Columns to focus on
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Salinity']
[76]:
# Look at important fields for max 5 values
salinity_series = df['Salinity'][df['Salinity'].notna()]
salinity_series.sort_values(ascending=False, inplace=True)
df[cols][df['Salinity'].isin(salinity_series[0:5])]
[76]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Salinity | |
|---|---|---|---|---|
| 14252 | 15030 | ppt | NaN | 15030.0 Practical_Salinity_Units |
| 24425 | 322 | ppth | NaN | 322.0 Practical_Salinity_Units |
| 68060 | 2150 | ppth | NaN | 2150.0 Practical_Salinity_Units |
| 100135 | 37782 | ppth | NaN | 37782.0 Practical_Salinity_Units |
| 170235 | 2190 | ppt | NaN | 2190.0 Practical_Salinity_Units |
Detection limits may help understand what caused low values and what results might need to be dropped or updated
[77]:
from harmonize_wq import wrangle
[78]:
df = wrangle.add_detection(df, 'Salinity')
cols+=['ResultDetectionConditionText',
'DetectionQuantitationLimitTypeName',
'DetectionQuantitationLimitMeasure/MeasureValue',
'DetectionQuantitationLimitMeasure/MeasureUnitCode']
[79]:
# Look at important fields for min 5 values (often multiple 0.0)
df[cols][df['Salinity'].isin(salinity_series[-5:])]
[79]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Salinity | ResultDetectionConditionText | DetectionQuantitationLimitTypeName | DetectionQuantitationLimitMeasure/MeasureValue | DetectionQuantitationLimitMeasure/MeasureUnitCode | |
|---|---|---|---|---|---|---|---|---|
| 1323 | 0 | ppt | NaN | 0.0 Practical_Salinity_Units | NaN | NaN | NaN | NaN |
| 2462 | 0.00 | ppth | NaN | 0.0 Practical_Salinity_Units | NaN | NaN | NaN | NaN |
| 3981 | 0 | ppt | NaN | 0.0 Practical_Salinity_Units | NaN | NaN | NaN | NaN |
| 4348 | 0.00 | ppth | NaN | 0.0 Practical_Salinity_Units | NaN | NaN | NaN | NaN |
| 4565 | 0 | ppt | NaN | 0.0 Practical_Salinity_Units | NaN | NaN | NaN | NaN |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 412123 | 0.0 | ppt | NaN | 0.0 Practical_Salinity_Units | NaN | NaN | NaN | NaN |
| 412344 | 0.0 | ppt | NaN | 0.0 Practical_Salinity_Units | NaN | NaN | NaN | NaN |
| 427637 | 0.0 | ppth | NaN | 0.0 Practical_Salinity_Units | NaN | NaN | NaN | NaN |
| 442693 | 0.0 | ppt | NaN | 0.0 Practical_Salinity_Units | NaN | NaN | NaN | NaN |
| 459763 | 0 | ppth | NaN | 0.0 Practical_Salinity_Units | NaN | NaN | NaN | NaN |
3051 rows × 8 columns
Explore Conductivity results:
[80]:
# Create series and inspect Conductivity values
cond_series = df['Conductivity'].dropna()
cond_series
[80]:
16 19204.2 microsiemens / centimeter
108 222.3 microsiemens / centimeter
218 102.8 microsiemens / centimeter
429 11017.5 microsiemens / centimeter
887 32.0 microsiemens / centimeter
...
463709 73.0 microsiemens / centimeter
463712 77.0 microsiemens / centimeter
463728 78.0 microsiemens / centimeter
463734 129.0 microsiemens / centimeter
463737 72.0 microsiemens / centimeter
Name: Conductivity, Length: 1818, dtype: object
Conductivity thresholds from Freshwater Explorer: 10 > x < 5000 us/cm, use a higher threshold for coastal waters
[81]:
# Sort and check other relevant columns before converting (e.g. Salinity)
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Salinity', 'Conductivity']
df.sort_values(by=['Conductivity'], ascending=False, inplace=True)
df.loc[df['Conductivity'].notna(), cols]
[81]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Salinity | Conductivity | |
|---|---|---|---|---|---|
| 155202 | 54886.2 | umho/cm | NaN | NaN | 54886.2 microsiemens / centimeter |
| 158906 | 54871.3 | umho/cm | NaN | NaN | 54871.3 microsiemens / centimeter |
| 151458 | 54860.6 | umho/cm | NaN | NaN | 54860.6 microsiemens / centimeter |
| 157517 | 54859.3 | umho/cm | NaN | NaN | 54859.3 microsiemens / centimeter |
| 150769 | 54850.8 | umho/cm | NaN | NaN | 54850.8 microsiemens / centimeter |
| ... | ... | ... | ... | ... | ... |
| 108539 | 6.8 | umho/cm | NaN | NaN | 6.8 microsiemens / centimeter |
| 170367 | 2 | umho/cm | NaN | NaN | 2.0 microsiemens / centimeter |
| 78435 | 2 | umho/cm | NaN | NaN | 2.0 microsiemens / centimeter |
| 30743 | 1 | umho/cm | NaN | NaN | 1.0 microsiemens / centimeter |
| 171757 | .04 | umho/cm | NaN | NaN | 0.04 microsiemens / centimeter |
1818 rows × 5 columns
[82]:
# Check other relevant columns before converting (e.g. Salinity)
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'QA_flag', 'Salinity', 'Conductivity']
df.loc[df['Conductivity'].notna(), cols]
[82]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | QA_flag | Salinity | Conductivity | |
|---|---|---|---|---|---|
| 155202 | 54886.2 | umho/cm | NaN | NaN | 54886.2 microsiemens / centimeter |
| 158906 | 54871.3 | umho/cm | NaN | NaN | 54871.3 microsiemens / centimeter |
| 151458 | 54860.6 | umho/cm | NaN | NaN | 54860.6 microsiemens / centimeter |
| 157517 | 54859.3 | umho/cm | NaN | NaN | 54859.3 microsiemens / centimeter |
| 150769 | 54850.8 | umho/cm | NaN | NaN | 54850.8 microsiemens / centimeter |
| ... | ... | ... | ... | ... | ... |
| 108539 | 6.8 | umho/cm | NaN | NaN | 6.8 microsiemens / centimeter |
| 170367 | 2 | umho/cm | NaN | NaN | 2.0 microsiemens / centimeter |
| 78435 | 2 | umho/cm | NaN | NaN | 2.0 microsiemens / centimeter |
| 30743 | 1 | umho/cm | NaN | NaN | 1.0 microsiemens / centimeter |
| 171757 | .04 | umho/cm | NaN | NaN | 0.04 microsiemens / centimeter |
1818 rows × 5 columns
[83]:
# Convert values to PSU and write to Salinity
cond_series = cond_series.apply(str) # Convert to string to convert to dimensionless (PSU)
df.loc[df['Conductivity'].notna(), 'Salinity'] = cond_series.apply(convert.conductivity_to_PSU)
df.loc[df['Conductivity'].notna(), 'Salinity']
[83]:
155202 36.356 dimensionless
158906 36.345 dimensionless
151458 36.338 dimensionless
157517 36.336 dimensionless
150769 36.33 dimensionless
...
108539 0.013 dimensionless
170367 0.012 dimensionless
78435 0.012 dimensionless
30743 0.012 dimensionless
171757 0.012 dimensionless
Name: Salinity, Length: 1818, dtype: object
Datetime
datetime() formats time using dataretrieval and ActivityStart
[84]:
# First inspect the existing unformated fields
cols = ['ActivityStartDate', 'ActivityStartTime/Time', 'ActivityStartTime/TimeZoneCode']
df[cols]
[84]:
| ActivityStartDate | ActivityStartTime/Time | ActivityStartTime/TimeZoneCode | |
|---|---|---|---|
| 155202 | 2007-08-09 | 12:15:00 | CST |
| 158906 | 2007-08-09 | 12:15:00 | CST |
| 151458 | 2007-08-09 | 12:15:00 | CST |
| 157517 | 2007-08-09 | 12:15:00 | CST |
| 150769 | 2007-08-09 | 12:15:00 | CST |
| ... | ... | ... | ... |
| 463839 | 1999-03-02 | 14:20:00 | CST |
| 463840 | 2001-11-28 | 12:05:00 | CST |
| 463841 | 2001-10-03 | 16:40:00 | CDT |
| 463842 | 2001-11-28 | 13:45:00 | CST |
| 463843 | 2001-10-03 | 14:15:00 | CDT |
463844 rows × 3 columns
[85]:
# 'ActivityStartDate' presserves date where 'Activity_datetime' is NAT due to no time zone
df = clean.datetime(df)
df[['ActivityStartDate', 'Activity_datetime']]
[85]:
| ActivityStartDate | Activity_datetime | |
|---|---|---|
| 155202 | 2007-08-09 | 2007-08-09 18:15:00+00:00 |
| 158906 | 2007-08-09 | 2007-08-09 18:15:00+00:00 |
| 151458 | 2007-08-09 | 2007-08-09 18:15:00+00:00 |
| 157517 | 2007-08-09 | 2007-08-09 18:15:00+00:00 |
| 150769 | 2007-08-09 | 2007-08-09 18:15:00+00:00 |
| ... | ... | ... |
| 463839 | 1999-03-02 | 1999-03-02 20:20:00+00:00 |
| 463840 | 2001-11-28 | 2001-11-28 18:05:00+00:00 |
| 463841 | 2001-10-03 | 2001-10-03 21:40:00+00:00 |
| 463842 | 2001-11-28 | 2001-11-28 19:45:00+00:00 |
| 463843 | 2001-10-03 | 2001-10-03 19:15:00+00:00 |
463844 rows × 2 columns
Activity_datetime combines all three time component columns into UTC. If time is missing this is NaT so a ActivityStartDate column is used to preserve date only.
Depth
Note: Data are often lacking sample depth metadata
[86]:
# Depth of sample (default units='meter')
df = clean.harmonize_depth(df)
#df.loc[df['ResultDepthHeightMeasure/MeasureValue'].dropna(), "Depth"]
df['ResultDepthHeightMeasure/MeasureValue'].dropna()
[86]:
1786 7.0
4191 7.0
79510 0.1
79597 2.2
79731 2.0
...
89816 2.2
110413 1.0
111001 16.0
111478 16.0
169725 35.0
Name: ResultDepthHeightMeasure/MeasureValue, Length: 179, dtype: float64
Characteristic to Column (long to wide format)
[87]:
# Split single QA column into multiple by characteristic (rename the result to preserve these QA_flags)
df2 = wrangle.split_col(df)
df2
[87]:
| OrganizationIdentifier | OrganizationFormalName | ActivityIdentifier | ActivityStartDate | ActivityStartTime/Time | ActivityStartTime/TimeZoneCode | MonitoringLocationIdentifier | ResultIdentifier | DataLoggerLine | ResultDetectionConditionText | ... | QA_TDP_Phosphorus | QA_Other_Phosphorus | QA_pH | QA_Carbon | QA_Conductivity | QA_Fecal_Coliform | QA_E_coli | QA_Salinity | QA_Turbidity | QA_Secchi | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 155202 | 21AWIC | ALABAMA DEPT. OF ENVIRONMENTAL MANAGEMENT - WA... | 21AWIC-51908_230231_173 | 2007-08-09 | 12:15:00 | CST | 21AWIC-1122 | STORET-170383613 | 230231.0 | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 158906 | 21AWIC | ALABAMA DEPT. OF ENVIRONMENTAL MANAGEMENT - WA... | 21AWIC-51908_230230_173 | 2007-08-09 | 12:15:00 | CST | 21AWIC-1122 | STORET-170383607 | 230230.0 | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 151458 | 21AWIC | ALABAMA DEPT. OF ENVIRONMENTAL MANAGEMENT - WA... | 21AWIC-51908_230228_173 | 2007-08-09 | 12:15:00 | CST | 21AWIC-1122 | STORET-170383595 | 230228.0 | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 157517 | 21AWIC | ALABAMA DEPT. OF ENVIRONMENTAL MANAGEMENT - WA... | 21AWIC-51908_230229_173 | 2007-08-09 | 12:15:00 | CST | 21AWIC-1122 | STORET-170383601 | 230229.0 | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 150769 | 21AWIC | ALABAMA DEPT. OF ENVIRONMENTAL MANAGEMENT - WA... | 21AWIC-51908_230227_173 | 2007-08-09 | 12:15:00 | CST | 21AWIC-1122 | STORET-170383589 | 230227.0 | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 463839 | USGS-AL | USGS Alabama Water Science Center | nwisal.01.99900500 | 1999-03-02 | 14:20:00 | CST | USGS-02376115 | NWIS-104002666 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 463840 | USGS-AL | USGS Alabama Water Science Center | nwisal.01.00201479 | 2001-11-28 | 12:05:00 | CST | USGS-02377570 | NWIS-53918846 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 463841 | USGS-AL | USGS Alabama Water Science Center | nwisal.01.00202076 | 2001-10-03 | 16:40:00 | CDT | USGS-02376115 | NWIS-104000948 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 463842 | USGS-AL | USGS Alabama Water Science Center | nwisal.01.00202072 | 2001-11-28 | 13:45:00 | CST | USGS-02376115 | NWIS-104000936 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 463843 | USGS-AL | USGS Alabama Water Science Center | nwisal.01.00201474 | 2001-10-03 | 14:15:00 | CDT | USGS-02377570 | NWIS-53918826 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
408983 rows × 117 columns
[88]:
# This expands the single col (QA_flag) out to a number of new columns based on the unique characteristicNames and speciation
print('{} new columns'.format(len(df2.columns) - len(df.columns)))
14 new columns
[89]:
# Note: there are fewer rows because NAN results are also dropped in this step
print('{} fewer rows'.format(len(df)-len(df2)))
54861 fewer rows
[90]:
#Examine Carbon flags from earlier in notebook (note these are empty now because NAN is dropped)
cols = ['ResultMeasureValue', 'ResultMeasure/MeasureUnitCode', 'Carbon', 'QA_Carbon']
df2.loc[df2['QA_Carbon'].notna(), cols]
[90]:
| ResultMeasureValue | ResultMeasure/MeasureUnitCode | Carbon | QA_Carbon |
|---|
Next the table is divided into the columns of interest (main_df) and characteristic specific metadata (chars_df)
[91]:
# split table into main and characteristics tables
main_df, chars_df = wrangle.split_table(df2)
[92]:
# Columns still in main table
main_df.columns
[92]:
Index(['OrganizationIdentifier', 'OrganizationFormalName',
'ActivityIdentifier', 'MonitoringLocationIdentifier', 'ProviderName',
'Secchi', 'Temperature', 'DO', 'pH', 'Salinity', 'Nitrogen',
'Speciation', 'TOTAL NITROGEN_ MIXED FORMS', 'Conductivity',
'Chlorophyll', 'Carbon', 'Turbidity', 'Sediment', 'Phosphorus',
'TP_Phosphorus', 'TDP_Phosphorus', 'Other_Phosphorus', 'Fecal_Coliform',
'E_coli', 'DetectionQuantitationLimitTypeName',
'DetectionQuantitationLimitMeasure/MeasureValue',
'DetectionQuantitationLimitMeasure/MeasureUnitCode',
'Activity_datetime', 'Depth', 'QA_Chlorophyll', 'QA_DO', 'QA_Nitrogen',
'QA_Temperature', 'QA_TP_Phosphorus', 'QA_TDP_Phosphorus',
'QA_Other_Phosphorus', 'QA_pH', 'QA_Carbon', 'QA_Conductivity',
'QA_Fecal_Coliform', 'QA_E_coli', 'QA_Salinity', 'QA_Turbidity',
'QA_Secchi'],
dtype='object')
[93]:
# look at main table results (first 5)
main_df.head()
[93]:
| OrganizationIdentifier | OrganizationFormalName | ActivityIdentifier | MonitoringLocationIdentifier | ProviderName | Secchi | Temperature | DO | pH | Salinity | ... | QA_TDP_Phosphorus | QA_Other_Phosphorus | QA_pH | QA_Carbon | QA_Conductivity | QA_Fecal_Coliform | QA_E_coli | QA_Salinity | QA_Turbidity | QA_Secchi | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 155202 | 21AWIC | ALABAMA DEPT. OF ENVIRONMENTAL MANAGEMENT - WA... | 21AWIC-51908_230231_173 | 21AWIC-1122 | STORET | NaN | NaN | NaN | NaN | 36.356 dimensionless | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 158906 | 21AWIC | ALABAMA DEPT. OF ENVIRONMENTAL MANAGEMENT - WA... | 21AWIC-51908_230230_173 | 21AWIC-1122 | STORET | NaN | NaN | NaN | NaN | 36.345 dimensionless | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 151458 | 21AWIC | ALABAMA DEPT. OF ENVIRONMENTAL MANAGEMENT - WA... | 21AWIC-51908_230228_173 | 21AWIC-1122 | STORET | NaN | NaN | NaN | NaN | 36.338 dimensionless | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 157517 | 21AWIC | ALABAMA DEPT. OF ENVIRONMENTAL MANAGEMENT - WA... | 21AWIC-51908_230229_173 | 21AWIC-1122 | STORET | NaN | NaN | NaN | NaN | 36.336 dimensionless | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 150769 | 21AWIC | ALABAMA DEPT. OF ENVIRONMENTAL MANAGEMENT - WA... | 21AWIC-51908_230227_173 | 21AWIC-1122 | STORET | NaN | NaN | NaN | NaN | 36.33 dimensionless | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
5 rows × 44 columns
[94]:
# Empty columns that could be dropped (Mostly QA columns)
cols = list(main_df.columns)
x = main_df.dropna(axis=1, how='all')
[col for col in cols if col not in x.columns]
[94]:
['Sediment',
'QA_TP_Phosphorus',
'QA_TDP_Phosphorus',
'QA_Other_Phosphorus',
'QA_Carbon',
'QA_Conductivity',
'QA_Fecal_Coliform',
'QA_E_coli',
'QA_Secchi']
[95]:
# Map average results at each station
gdf_avg = visualize.map_measure(main_df, stations_clipped, 'Temperature')
gdf_avg.plot(column='mean', cmap='OrRd', legend=True)
[95]:
<Axes: >