3 Model Fitting
Throughout this section, we will use both the spmodel package and the ggplot2 package:
Goals:
- Further explore the
splm()function using themossdata. - Connect parameter estimates in the summary output of
splm()to the spatial linear model.
3.1 Data Introduction
The moss data in the spmodel package is an sf (simple features) object (Pebesma 2018) that contains observations on heavy metals in mosses near a mining road in Alaska. An sf object is a special data.frame built for storing spatial information and contains a column called geometry. We can view the first few rows of moss by running
moss
#> Simple feature collection with 365 features and 7 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: -445884.1 ymin: 1929616 xmax: -383656.8 ymax: 2061414
#> Projected CRS: NAD83 / Alaska Albers
#> # A tibble: 365 × 8
#> sample field_dup lab_rep year sideroad log_dist2road log_Zn
#> <fct> <fct> <fct> <fct> <fct> <dbl> <dbl>
#> 1 001PR 1 1 2001 N 2.68 7.33
#> 2 001PR 1 2 2001 N 2.68 7.38
#> 3 002PR 1 1 2001 N 2.54 7.58
#> 4 003PR 1 1 2001 N 2.97 7.63
#> 5 004PR 1 1 2001 N 2.72 7.26
#> 6 005PR 1 1 2001 N 2.76 7.65
#> # ℹ 359 more rows
#> # ℹ 1 more variable: geometry <POINT [m]>More information about moss can be found by running help("moss", "spmodel").
Our goal is to model the distribution of log zinc concentration (log_Zn) using a spatial linear model. We can visualize the distribution of log zinc concentration (log_Zn) in moss by running
ggplot(moss, aes(color = log_Zn)) +
geom_sf(size = 2) +
scale_color_viridis_c() +
scale_x_continuous(breaks = seq(-163, -164, length.out = 2)) +
theme_gray(base_size = 14)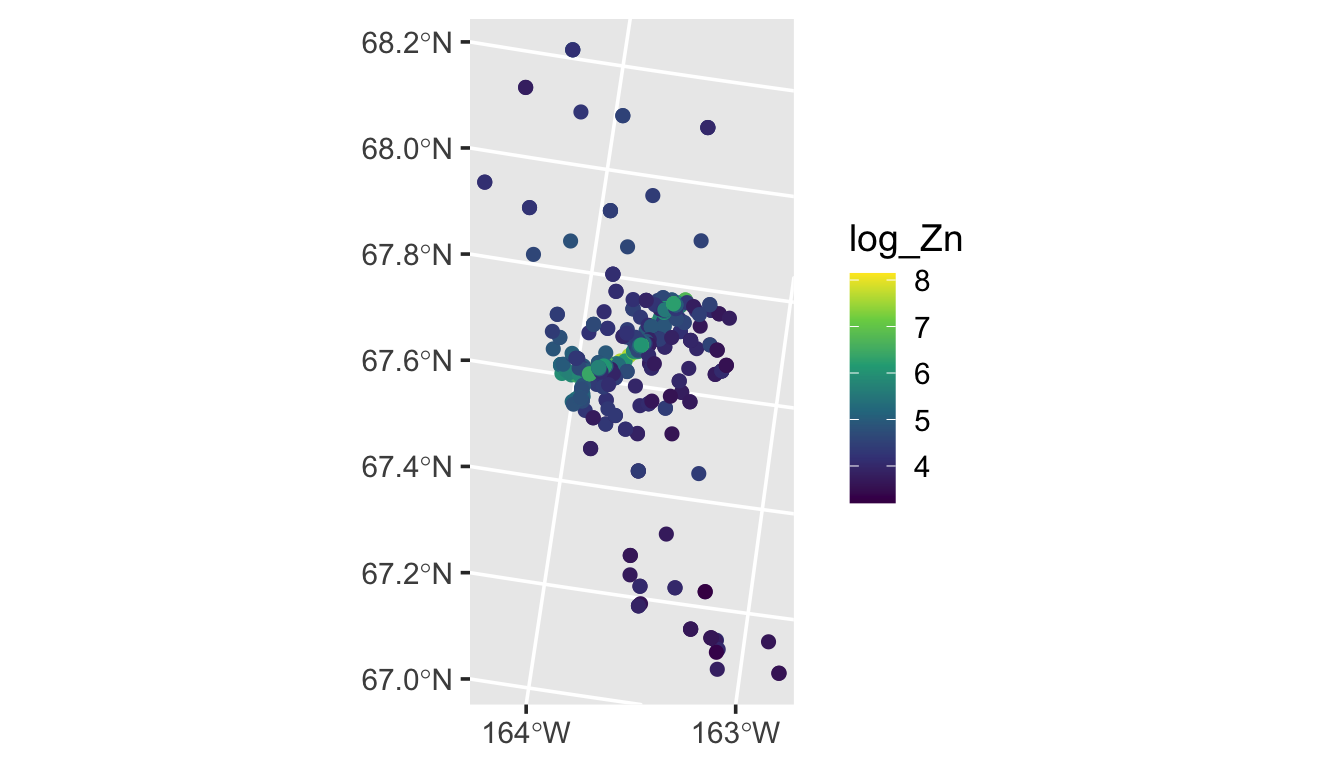
An important predictor variable may be the log of the distance to the haul road, log_dist2road, which is measured in meters. Next we use spmodel to fit a spatial linear model with with log_Zn as the response and log_dist2road as a predictor.
3.2 splm() Syntax and Output Interpretation
The splm() function shares similar syntactic structure with the lm() function used to fit linear models without spatial dependence (Equation 2.1). splm() generally requires at least three arguments
-
formula: a formula that describes the relationship between the response variable (\(\mathbf{y}\)) and explanatory variables (\(\mathbf{X}\)) -
data: adata.frameorsfobject that contains the response variable, explanatory variables, and spatial information. -
spcov_type: the spatial covariance type ("exponential","matern","spherical", etc)- There are 17 different types
If data is an sf object, then spatial information is stored in the object’s geometry. However, if data is a data.frame or tibble (a special data.frame), then the names of the variables that represent the x-coordinates and y-coordinates must also be provided as two additional arguments via xcoord and ycoord.
We fit a spatial linear model regressing log zinc concentration (log_Zn) on log distance to a haul road (log_dist2road) using an exponential spatial covariance function by running
spmod <- splm(formula = log_Zn ~ log_dist2road, data = moss,
spcov_type = "exponential")The estimation method in splm() is specified by estmethod. The default estimation method is restricted maximum likelihood ("reml"). Additional options include maximum likelihood "ml", semivariogram-based composite likelihood ("sv-cl") (Curriero and Lele 1999), and semivariogram-based weighted least squares ("sv-wls") (Cressie 1985). When the estimation method is semivariogram-based weighted least squares, the weights are specified by weights with a default of Cressie weights (“cressie").
We summarize the model fit by running
summary(spmod)
#>
#> Call:
#> splm(formula = log_Zn ~ log_dist2road, data = moss, spcov_type = "exponential")
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -2.6801 -1.3606 -0.8103 -0.2485 1.1298
#>
#> Coefficients (fixed):
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 9.76825 0.25216 38.74 <2e-16 ***
#> log_dist2road -0.56287 0.02013 -27.96 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Pseudo R-squared: 0.683
#>
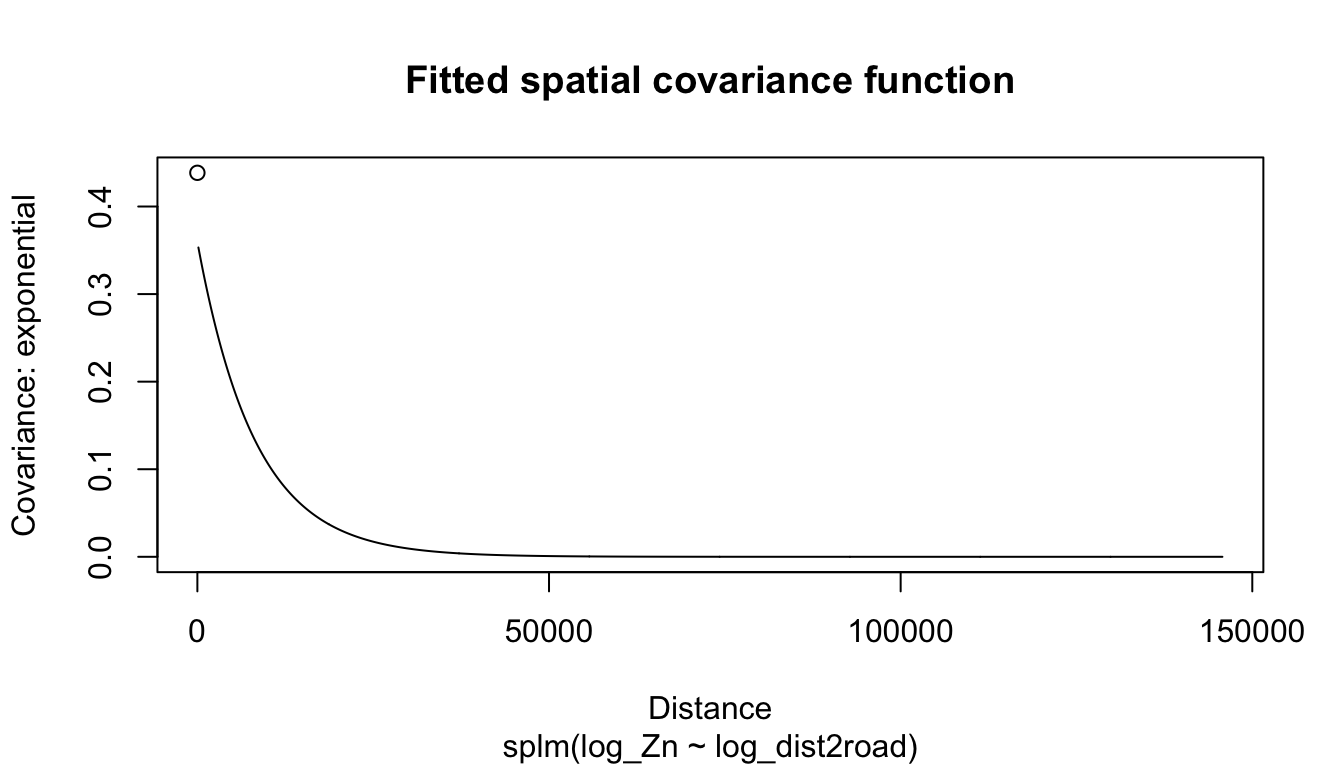
#> Coefficients (exponential spatial covariance):
#> de ie range
#> 3.595e-01 7.897e-02 8.237e+03The fixed effects coefficient table contains estimates, standard errors, z-statistics, and asymptotic p-values for each fixed effect. From this table, we notice there is evidence that mean log zinc concentration significantly decreases with distance from the haul road (p-value < 2e-16).
We can relate some of the components in the summary output to the model in Equation 2.2:
- The values in the
Estimatecolumn of theCoefficients (fixed)table form \(\boldsymbol{\hat{\beta}}\), an estimate of \(\boldsymbol{\beta}\). - The
devalue of 0.36 in theCoefficients (exponential spatial covariance)table is \(\hat{\sigma}^2_{de}\), which is an estimate of \(\sigma^2_{de}\), the variance of \(\boldsymbol{\tau}\) (commonly called the partial sill). - The
ievalue of 0.079 in theCoefficients (exponential spatial covariance)table is \(\hat{\sigma}^2_{ie}\), which is an estimate of \(\sigma^2_{ie}\), the variance of \(\boldsymbol{\epsilon}\) (commonly called the nugget). - The
rangevalue of 8,237 in theCoefficients (exponential spatial covariance)table is \(\hat{\phi}\), which is an estimate of \(\phi\) (recall \(\phi\) is the range parameter in Equation 2.3 that controls the behavior of the spatial covariance as a function of distance).
The summary() output, while useful, is printed to the R console and not easy to manipulate. The tidy() function turns the coefficient table into a tibble (i.e., a special data.frame) that is easy to manipulate. We tidy the fixed effects by running
tidy(spmod)
#> # A tibble: 2 × 5
#> term estimate std.error statistic p.value
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 (Intercept) 9.77 0.252 38.7 0
#> 2 log_dist2road -0.563 0.0201 -28.0 0We tidy the spatial covariance parameters by running
tidy(spmod, effects = "spcov")
#> # A tibble: 3 × 3
#> term estimate is_known
#> <chr> <dbl> <lgl>
#> 1 de 0.360 FALSE
#> 2 ie 0.0790 FALSE
#> 3 range 8237. FALSEThe is_known column indicates whether the parameter is assumed known. By default, all parameters are assumed unknown. We discuss this more in Chapter 5.
3.3 Model Fit and Diagnostics
The quality of model fit can be assessed using a variety of statistics readily available in spmodel, including AIC, AICc, and pseudo R-squared. Additionally, model diagnostics such as leverage, fitted values, residuals (several types), and Cook’s distance. While both the model fit statistics and the diagnostics can be found with individual functions like AIC(), residuals(), cooks.distance(), etc., they can also be computed using glance() (for the model fit statistics) and augment() (for the diagnostics).
glance(spmod)
#> # A tibble: 1 × 9
#> n p npar value AIC AICc logLik deviance pseudo.r.squared
#> <int> <dbl> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 365 2 3 367. 373. 373. -184. 363 0.683The output from glance() shows model fit statistics for the spatial linear model with an exponential covariance structure for the errors.
The augment() function provides many model diagnostics statistics in a single tibble:
augment(spmod)
#> Simple feature collection with 365 features and 7 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: -445884.1 ymin: 1929616 xmax: -383656.8 ymax: 2061414
#> Projected CRS: NAD83 / Alaska Albers
#> # A tibble: 365 × 8
#> log_Zn log_dist2road .fitted .resid .hat .cooksd .std.resid
#> * <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 7.33 2.68 8.26 -0.928 0.0200 0.0142 1.18
#> 2 7.38 2.68 8.26 -0.880 0.0200 0.0186 1.35
#> 3 7.58 2.54 8.34 -0.755 0.0225 0.00482 0.647
#> 4 7.63 2.97 8.09 -0.464 0.0197 0.0305 1.74
#> 5 7.26 2.72 8.24 -0.977 0.0215 0.131 3.45
#> 6 7.65 2.76 8.21 -0.568 0.0284 0.0521 1.89
#> # ℹ 359 more rows
#> # ℹ 1 more variable: geometry <POINT [m]>augment() returns a tibble with many model diagnostics statistics, including
-
.fitted, the fitted value, calculated from the estimated fixed effects in the model -
.hat, the Mahalanobis distance, a metric of leverage -
.cooksd, the Cook’s distance, a metric of influence -
.std.resid, the standardized residual
If the model is correct, then the standardized residuals have mean 0, standard deviation 1, and are uncorrelated.
The plot() function can be used on a fitted model object to construct a few pre-specified plots of these model diagnostics. For example, the following code plots the Cook’s distance, a measure of influence, which quantifies each observation’s impact on model fit:
plot(spmod, which = 4)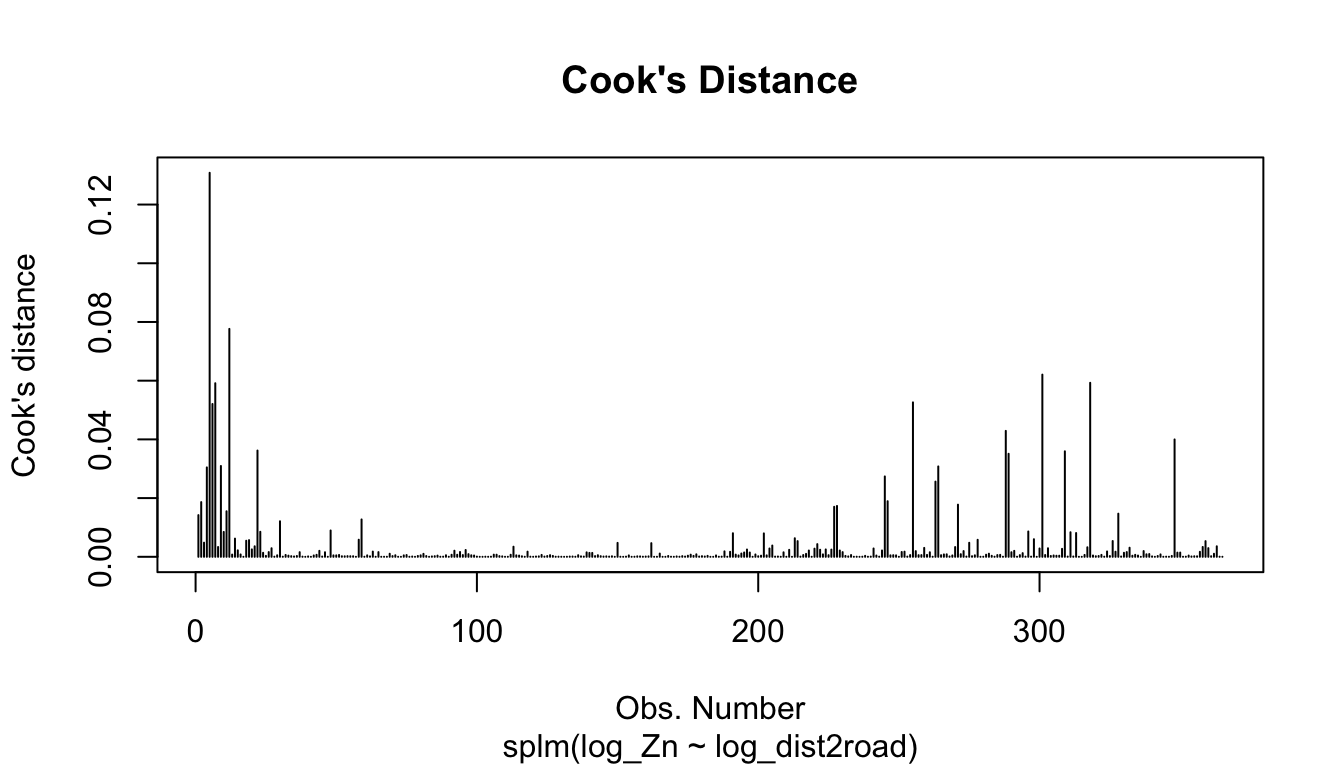
The other 7 plots for model objects fit with splm() can be read about in the help: ?plot.spmodel.
If the grammar of graphics plotting syntax in ggplot2 is more familiar, then we can also construct plots with the augmented model:
aug_df <- augment(spmod)
ggplot(data = aug_df, aes(x = seq_len(nrow(aug_df)),
y = .cooksd)) +
geom_point() +
theme_minimal() +
labs(x = "Row Number")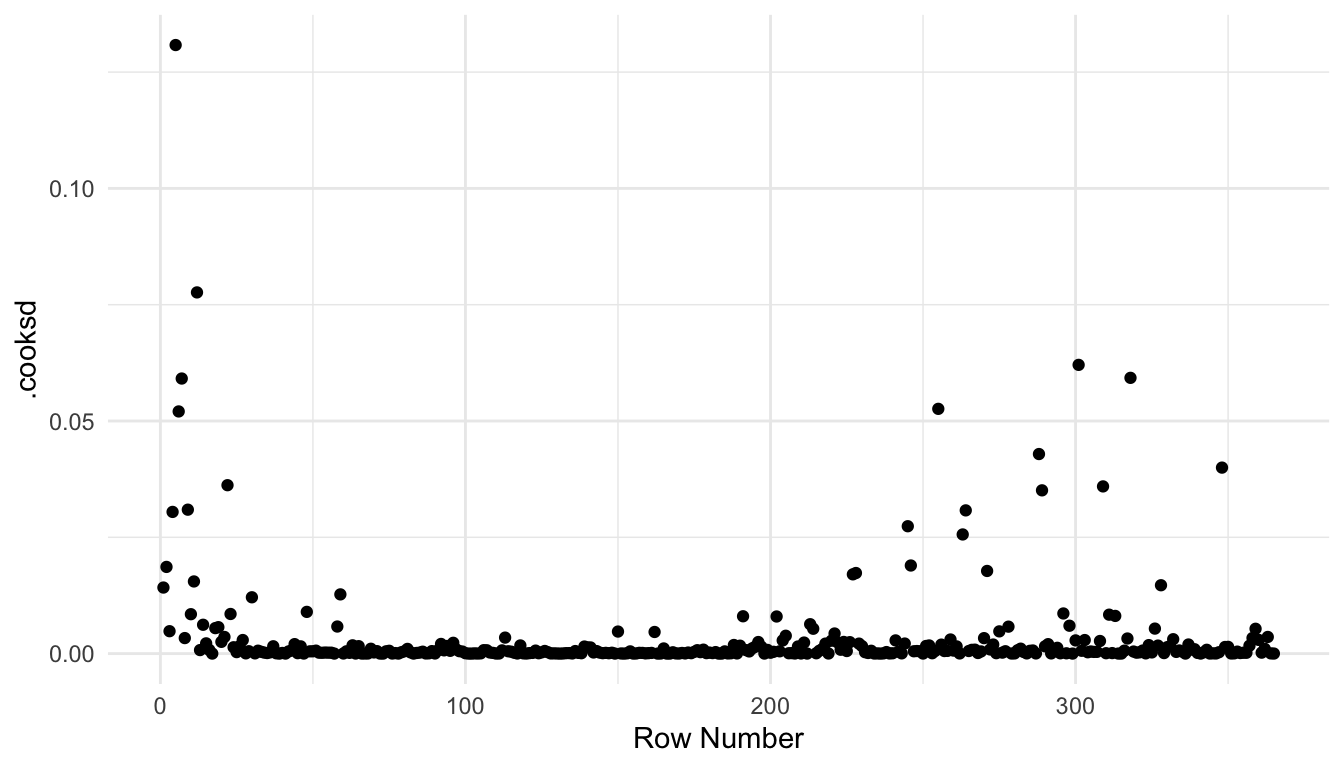
3.4 R Code Appendix
library(spmodel)
library(ggplot2)
moss
ggplot(moss, aes(color = log_Zn)) +
geom_sf(size = 2) +
scale_color_viridis_c() +
scale_x_continuous(breaks = seq(-163, -164, length.out = 2)) +
theme_gray(base_size = 14)
spmod <- splm(formula = log_Zn ~ log_dist2road, data = moss,
spcov_type = "exponential")
summary(spmod)
tidy(spmod)
tidy(spmod, effects = "spcov")
caribou_mod <- splm(z ~ tarp + water + tarp:water,
data = caribou, spcov_type = "pexponential",
xcoord = x, ycoord = y)
summary(caribou_mod)
anova(caribou_mod)
tidy(anova(caribou_mod))
glance(spmod)
mat <- splm(formula = log_Zn ~ log_dist2road, data = moss,
spcov_type = "matern")
none <- splm(formula = log_Zn ~ log_dist2road, data = moss,
spcov_type = "none")
glances(spmod, mat, none)
augment(spmod)
plot(spmod, which = 4)
aug_df <- augment(spmod)
ggplot(data = aug_df, aes(x = seq_len(nrow(aug_df)),
y = .cooksd)) +
geom_point() +
theme_minimal() +
labs(x = "Row Number")
plot(spmod, which = 7)