Spatial Generalized Linear Models in spmodel
Michael Dumelle, Matt Higham, and Jay M. Ver Hoef
Source:vignettes/articles/SPGLMs.Rmd
SPGLMs.RmdIntroduction
spmodel is an R package used to fit,
summarize, and predict for a variety of spatial statistical models. The
vignette provides an introduction spatial generalized linear models for
non-Gaussian response distributions in spmodel. Before
proceeding, we load spmodel by running
If using spmodel in a formal publication or report,
please cite it. Citing spmodel lets us devote more
resources to the package in the future. We view the spmodel
citation by running
citation(package = "spmodel")#> To cite spmodel in publications use:
#>
#> Dumelle M, Higham M, Ver Hoef JM (2023). spmodel: Spatial statistical
#> modeling and prediction in R. PLOS ONE 18(3): e0282524.
#> https://doi.org/10.1371/journal.pone.0282524
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Article{,
#> title = {{spmodel}: Spatial statistical modeling and prediction in {R}},
#> author = {Michael Dumelle and Matt Higham and Jay M. {Ver Hoef}},
#> journal = {PLOS ONE},
#> year = {2023},
#> volume = {18},
#> number = {3},
#> pages = {1--32},
#> doi = {10.1371/journal.pone.0282524},
#> url = {https://doi.org/10.1371/journal.pone.0282524},
#> }We will create visualizations using ggplot2 (Wickham 2016), which we load by running
ggplot2 is only installed alongside spmodel
when dependencies = TRUE in
install.packages(), so check that the package is installed
and loaded before reproducing any of these vignette’s visualizations. We
will also show code that can be used to create interactive
visualizations of spatial data with mapview (Appelhans et al. 2022). mapview
also has many backgrounds available that contextualize spatial data with
topographical information. Before running the mapview code
interactively, make sure mapview is installed and
loaded.
spmodel contains various methods for generic functions
defined outside of spmodel. To find relevant documentation
for these methods, run help("generic.spmodel", "spmodel")
(e.g., help("summary.spmodel", "spmodel"),
help("predict.spmodel", "spmodel"), etc.). Note that
?generic.spmodel is shorthand for
help("generic.spmodel", "spmodel"). We provide more details
and examples regarding these methods and generics throughout this
vignette. For a full list of spmodel functions available,
see spmodel’s documentation manual.
A Review of the Spatial Linear Model
We first focus on spatial linear models for a Gaussian response vector. The spatial linear model is defined as \[\begin{equation}\label{eq:splm} \mathbf{y} = \mathbf{X} \boldsymbol{\beta} + \boldsymbol{\tau} + \boldsymbol{\epsilon}, \end{equation}\] where for a sample size \(n\), \(\mathbf{y}\) is an \(n \times 1\) column vector of response variables, \(\mathbf{X}\) is an \(n \times p\) design (model) matrix of explanatory variables, \(\boldsymbol{\beta}\) is a \(p \times 1\) column vector of fixed effects controlling the impact of \(\mathbf{X}\) on \(\mathbf{y}\), \(\boldsymbol{\tau}\) is an \(n \times 1\) column vector of spatially dependent random errors, and \(\boldsymbol{\epsilon}\) is an \(n \times 1\) column vector of spatially independent random errors. We make a few assumptions about \(\boldsymbol{\tau}\) and \(\boldsymbol{\epsilon}\) in the spatial linear model: first, that \(\text{E}(\boldsymbol{\tau}) = \text{E}(\boldsymbol{\epsilon}) = \boldsymbol{0}\), where \(\text{E}(\cdot)\) denotes expectation; second, that \(\text{Cov}(\boldsymbol{\tau}) = \sigma^2_\tau \mathbf{R}\), where \(\mathbf{R}\) is an \(n \times n\) matrix that determines the spatial dependence structure in \(\mathbf{y}\) and depends on a range parameter, \(\phi\); third, that \(\text{Cov}(\boldsymbol{\epsilon}) = \sigma^2_\epsilon \mathbf{I}\), where \(\mathbf{I}\) is an \(n \times n\) identity matrix; and fourth, that \(\boldsymbol{\tau}\) and \(\boldsymbol{\epsilon}\) are independent of one another. The parameter \(\sigma^2_{\tau}\) is called the spatially dependent random error variance or partial sill. The parameter \(\sigma^2_\epsilon\) is called the spatially independent random error variance or nugget. These two variance parameters are henceforth more intuitively written as \(\sigma^2_{de}\) and \(\sigma^2_{ie}\), respectively. The covariance of \(\mathbf{y}\) is denoted \(\boldsymbol{\Sigma}\) and given by \[\begin{equation}\label{eq:spcov} \boldsymbol{\Sigma} = \sigma^2_{de}\mathbf{R} + \sigma^2_{ie} \mathbf{I}. \end{equation}\] The parameters \(\sigma^2_{de}\), \(\phi\), and \(\sigma^2_{ie}\) are elements of \(\boldsymbol{\theta}\), the covariance parameter vector.
The spatial linear model applies to both point-referenced and areal
(i.e., lattice; polygon) data. Spatial data are point-referenced when
the elements in \(\mathbf{y}\) are
observed at point-locations indexed by x-coordinates and y-coordinates
on a spatially continuous surface with an infinite number of locations.
For example, consider sampling soil at any point-location in a field.
Spatial linear models for point-referenced data are sometimes called
geostatistical models. An example of an \(\mathbf{R}\) matrix for point-referenced
data is the spherical correlation matrix given by \[\begin{equation}\label{eq:spherical}
\mathbf{R} = \left(1 - 1.5\frac{\mathbf{H}}{\phi} +
0.5\frac{\mathbf{H}^3}{\phi^3}\right) \odot \mathcal{I}(\mathbf{H} \leq
\phi),
\end{equation}\]
where \(\mathbf{H}\) is a matrix of
Euclidean distances among observations, \(\mathbf{H}^3\) is a matrix of cubed
Euclidean distances, \(\odot\) is the
Hadmard (element-wise; direct) product, and \(\mathcal{I}(\mathbf{H} \leq \phi)\) is an
indicator function equal to one when the \(ij\)th element of \(\mathbf{H}\) is less than \(\phi\) (the range) and zero otherwise. A
second is example is the Matérn correlation matrix given by \[\begin{equation}\label{eq:matern}
\mathbf{R} = \frac{2^{1 - \nu}}{\Gamma(\nu)} \left(2\nu \mathbf{H} /
\phi \right)^{\nu / 2} \odot B_k(2\nu \mathbf{H}, \nu),
\end{equation}\] where \(\nu\)
is a smoothness parameter, \(\odot\) is
the Hadmart (element-wise; direct) product, and \(B_k(\cdot, \nu)\) is a Bessel-K function
with order \(\nu\). Chiles and Delfiner (2012) provide examples of
several spatial dependence structures.
Spatial data are areal when they are observed as part of a finite network of polygons whose connections are indexed by a neighborhood structure. For example, the polygons may represent counties in a state who are neighbors if they share at least one boundary. An example of an \(\mathbf{R}\) matrix for areal data is the simultaneous-autoregressive spatial correlation matrix given by \[\begin{equation*} \mathbf{R} = [(\mathbf{I} - \phi \mathbf{W})(\mathbf{I} - \phi \mathbf{W})^\top]^{-1}, \end{equation*}\] where \(\mathbf{W}\) is a weight matrix that describes the neighborhood structure among observations. Spatial linear models for areal data are sometimes called spatial autoregressive models. For thorough reviews of the spatial linear model, see Cressie (1993), Banerjee, Carlin, and Gelfand (2014), and Schabenberger and Gotway (2017).
The Spatial Generalized Linear Model
It is common to implicitly assume \(\mathbf{y}\) in the spatial linear model is unconstrained and Gaussian. This is a restrictive assumption and often inappropriate for binary data, proportion data, count data, and skewed data, which are common in many practical applications. These types of data are most naturally described using generalized linear models (Nelder and Wedderburn 1972; McCullagh and Nelder 1989). Generalized linear models are an extension of linear models that “link” a function of a distribution’s mean to the linear function \(\mathbf{X} \boldsymbol{\beta}\) (the explanatory variables and fixed effects). We can extend generalized linear models by adding linear, latent (i.e., unobserved) random effects to \(\mathbf{X} \boldsymbol{\beta}\). These types of models are called generalized linear mixed models (Breslow and Clayton 1993) and are often formulated using a hierarchical construction (Lee and Nelder 1996). For thorough reviews of generalized linear models and generalized linear mixed models, see Bolker et al. (2009), Myers et al. (2012), Faraway (2016), and Wood (2017).
A recent spmodel release (version 0.4.0) provided
support for spatial generalized linear models (i.e., a generalized
linear mixed model with spatial random effects). We define the spatial
generalized linear model as \[\begin{equation}\label{eq:spglm}
f(\boldsymbol{\mu}) \equiv \mathbf{w} = \mathbf{X} \boldsymbol{\beta} +
\boldsymbol{\tau} + \boldsymbol{\epsilon},
\end{equation}\] where \(f(\cdot)\) is the link function, \(\boldsymbol{\mu}\) is the mean of \(\mathbf{y}\), and the remaining terms \(\mathbf{X}\), \(\boldsymbol{\beta}\), \(\boldsymbol{\tau}\), \(\boldsymbol{\epsilon}\) represent the same
quantities as for spatial linear models. That is, \(\text{E}(\boldsymbol{\tau}) =
\text{E}(\boldsymbol{\epsilon}) = \boldsymbol{0}\), \(\text{Cov}(\boldsymbol{\tau}) =
\sigma^2_{de}\mathbf{R}\), \(\text{Cov}(\boldsymbol{\epsilon}) = \sigma^2_{ie}
\mathbf{I}\), \(\boldsymbol{\tau}\) is independent of \(\boldsymbol{\epsilon}\), and \(\boldsymbol{\Sigma} = \sigma^2_{de} \mathbf{R} +
\sigma^2_{ie} \mathbf{I}\). The link function, \(f(\cdot)\), “links” a function of \(\boldsymbol{\mu}\) to the linear term \(\mathbf{w}\). Note that the linking of
\(\boldsymbol{\mu}\) to \(\mathbf{w}\) applies element-wise to each
vector. Each link function \(f(\cdot)\)
has a corresponding inverse link function, \(f^{-1}(\cdot)\), which “links” a function
of \(\mathbf{w}\) to \(\boldsymbol{\mu}\). Notice that \(\mathbf{w}\) is unconstrained but \(\boldsymbol{\mu}\) is usually constrained
in some way (e.g., positive) that depends on the distribution assumed
for \(\mathbf{y}\).
Spatial linear and generalized linear models rely on the same components: explanatory variables, fixed effects, spatially dependent random errors, and spatial independent random errors. These components are related to the response linearly (spatial linear models) or non-linearly (spatial generalized linear models). The response itself is generally assumed to follow a Gaussian distribution (spatial linear models) or one of many non-Gaussian distributions (spatial generalized linear models). Some non-Gaussian distributions include the binomial distribution for binary responses, the beta distribution for proportion responses, the Poisson or negative binomial distributions for count responses, and the gamma or inverse Gaussian distributions for skewed, positive continuous responses.
spmodel implements a marginal approach to inference that
uses a novel application of the Laplace approximation. This marginal
approach is powerful and flexible, accommodating a wide range of
covariance structures. It is a frequentist approach, which means that it
does not rely on any Bayesian sampling routines, as is common when
handling complex hierarchical structures. It also formally maximizes a
likelihood, which is advantageous because it means that likelihood-based
statistics such as AIC (Akaike 1974; Hoeting et
al. 2006) and BIC (Schwarz 1978)
are defined, in contrast to approaches that only specify the first two
moments of a distribution (e.g., quasi-likelihood fit via iteratively
re-weighted least squares). Ver Hoef et al.
(2024) provide the methodology’s full details.
spmodel accommodates the binomial, beta, Poisson,
negative binomial, gamma, and inverse Gaussian distributions. The
binomial and beta response distributions (specified by
family) use the logit link, while the Poisson, negative
binomial, gamma, and inverse Gaussian distributions use the log link –
see the following table.
| Family | Link Function | Link Name | Data Type |
|---|---|---|---|
| Binomial | \(f(\mathbf{\mu}) = \log(\mathbf{\mu} / (1 - \mathbf{\mu}))\) | Logit | Binary; Binary Count |
| Poisson | \(f(\mathbf{\mu}) = \log(\mathbf{\mu})\) | Log | Count |
| Negative Binomial | \(f(\mathbf{\mu}) = \log(\mathbf{\mu})\) | Log | Count |
| Beta | \(f(\mathbf{\mu}) = \log(\mathbf{\mu} / (1 - \mathbf{\mu}))\) | Logit | Proportion |
| Gamma | \(f(\mathbf{\mu}) = \log(\mathbf{\mu})\) | Log | Skewed |
| Inverse Gaussian | \(f(\mathbf{\mu}) = \log(\mathbf{\mu})\) | Log | Skewed |
Model fitting
Spatial generalized linear models in spmodel are fit
using spglm() (for point-referenced data) or
spgautor() (for areal data). These functions are similar in
structure both to one another and to the glm() function in
base R. They generally require the following four
arguments: formula, a formula that describes the
relationship between the response and explanatory variables;
family, the response distribution, which can be
binomial, beta, poisson,
nbinomial, Gamma, or
inverse.gaussian; data, a
data.frame or sf object (Pebesma 2018) whose rows index observations and
whose columns contain the response variable, explanatory variables, and
spatial information; and spcov_type, one of 19 spatial
covariance types (e.g., spherical spatial covariance, Matérn spatial
covariance, simultaneous-autoregressive spatial covariance etc.). The
binomial and beta response distributions (specified by
family) use the logit link, while the Poisson, negative
binomial, gamma, and inverse Gaussian distributions use the log link.
Other arguments to spglm() and spgautor() are
either required only in specific situations or have default values
specified. All arguments to the spglm() and
spgautor() functions are summarized in the following table.
Models for several hundred observations can take anywhere from a few
seconds to a few minutes to fit, depending on the shape of the
likelihood surface and how quickly the covariance parameters converge.
Models for several thousand observations can utilize
spmodel’s large data set options.
| Argument | Purposes | Applies To |
|---|---|---|
formula |
Fixed effects formula | Both |
family |
Response distribution | Both |
data |
Data Frame or sf object |
Both |
spcov_type |
Spatial covariance type | Both |
spcov_initial |
Initial or known spatial covariance parameters | Both |
dispersion_initial |
Initial or known dispersion parameter | Both |
estmethod |
Estimation method | Both |
random |
Random effects formula | Both |
randcov_initial |
Initial or known random effect variances | Both |
partition_factor |
A partition factor formula | Both |
... |
Additional arguments | Both |
anisotropy |
Whether to model anisotropy | spglm() |
xcoord |
An x-coordinate name | spglm()} |
ycoord |
A y-coordinate name | spglm() |
local |
Options for large data sets | spglm() |
W |
A neighborhood weight matrix | spgautor() |
row_st |
Whether to row-standardize W
|
spgautor() |
M |
A symmetry matrix | spgautor() |
range_positive |
Whether the range parameter is restricted to be positive | |
cutoff |
Distance cutoff if using distance-based neighbor definition |
Model evaluation and diagnostics
After fitting a model, it is usually beneficial to evaluate certain
aspects of model fit or inspect and visualize model diagnostics like
residuals. There are many functions in spmodel that are
used for these purposes, and their main argument is the fitted model
object returned by spglm() or spgautor(). The
names of these functions are meant to be illustrative, describing the
purpose of the function itself. The functions for evaluating model fit
(see below) return likelihood-based statistics like AIC, AICc, BIC, and
deviance (McCullagh and Nelder 1989),
return coefficient estimates and confidence intervals, perform cross
validation, and more.
| Function | Purpose |
|---|---|
AIC(); AICc()
|
Return AIC and AICc |
anova() |
Perform an analysis of variance |
AUROC() |
Compute the area under the receiver operating characteristic curve |
BIC() |
Return BIC |
coef(); coefficients()
|
Return parameter estimates |
confint() |
Return confidence intervals |
covmatrix() |
Return the fitted covariance matrix |
deviance() |
Return the deviance |
glance(); glances()
|
Glance at model fits |
logLik() |
Return the log-likelihood |
loocv() |
Perform leave-one-out cross validation |
pseudoR2() |
Return the pseudo R-squared |
summary() |
Summarize the fitted model |
tidy() |
Tidy the fitted model |
varcomp() |
Compare sources of variability |
vcov() |
Return variance-covariance matrices |
The functions for inspecting model diagnostics (see below) return Cook’s distances (Cook 1979), fitted values, leverage (hat) values, residuals (of several types), and visualizations.
| Function | Purpose |
|---|---|
augment() |
Augment model data with diagnostics |
cooks.distance() |
Return Cook’s distances |
fitted(); fitted.values()
|
Return fitted values |
hatvalues(); |
Return hat (leverage) values |
plot() |
Plot model diagnostics |
residuals(); resid();
rstandard()
|
Return residuals |
The functions tidy(), glance(), and
augment(), popularized by the broom
R package (Robinson, Hayes, and
Couch 2021), are particularly useful. The tidy()
function tidies parameter estimate output, returning a special
data.frame called a tibble (Müller and Wickham 2021) that is much easier to
manipulate and work with than the parameter estimate output printed by
summary() to the R console. The
glance() function glances at the model fit, returning a
tibble with the sample size, number of estimated
parameters, and several model-fit statistics. glances() is
an extension of glance() that glances at multiple models
simultaneously. The augment() function augments the model
fit, returning a tibble with the data used to fit the model
as well as model diagnostics. The augment() function can
also be used to augment prediction data, as we discuss next.
Prediction
Often a priority of a spatial data analysis is using the fitted model
to make predictions at unobserved locations and quantify uncertainties
in those predictions. The predict() function in
spmodel is used to predict the mean process at unobserved
locations. It is similar in structure to predict() for
objects fit using glm() in base R and
takes two main arguments: object, the fitted model object
returned by spglm() or spgautor(); and
newdata, a data.frame or sf
object whose rows index locations requiring prediction and whose columns
contain the explanatory variables and spatial information at these
locations. Other arguments to predict() are either required
only in specific situations or have default values specified. All
arguments to the predict() function are summarized in the
following table. The augment() function can also be used to
augment newdata with predictions, taking the same arguments
as predict().
| Argument | Purpose |
|---|---|
object |
Fitted model object |
newdata |
New data frame or sf object |
type |
Whether predictions are on the link or response scale |
se.fit |
Whether standard errors should be returned |
interval |
Interval type (prediction or confidence) |
level |
Prediction or confidence level |
local |
Options for large data sets |
var_correct |
Whether prediction variances should be adjusted |
... |
Additional arguments |
Simulation
spmodel has functions that are used to simulate spatial
data from Gaussian, binomial, beta, Poisson, negative binomial, gamma,
and inverse Gaussian response distributions. These simulation functions
are similar in structure to commonly used base R
functions for simulating data. They are also similar in name, simply
adding an sp prefix to the conventional name in base
R. For example, rpois() is used to
simulate Poisson data in base R, while
sprpois() is used to simulate spatial Poisson data in
spmodel. These functions are summarized in the following
table.
| Function | Purpose |
|---|---|
sprnorm() |
Simulate spatial Gaussian data |
sprbinom() |
Simulate spatial binomial data |
sprbeta() |
Simulate spatial beta data |
sprpois() |
Simulate spatial Poisson data |
sprnbinom() |
Simulate spatial negative binomial data |
sprgamma() |
Simulate spatial gamma data |
sprinvgauss() |
Simulate spatial inverse Gaussian data |
An application to binary (presence/absence) data
In the next two sections we to show how to use spmodel
to analyze data collected on moose in the Togiak region of Alaska, USA.
The moose data in spmodel contain information
on moose counts from a survey of sites in the Togiak region. The data
are stored as an sf object, a special
data.frame built to store spatial data. moose
has several variables: strat; a stratification variable
with two levels (L for low and M for medium)
based on surrounding landscape metrics; elev, the site
elevation; presence, whether or not at least one moose was
observed at the site; count, the number of moose observed
at a the site; and geometry, the spatial coordinates in an
Alaska Albers projection (EPSG: 3338). We load spmodel and
the moose data by running
We are interested in quantifying the effects of elev and
strat on moose presence and begin with some
visualizations. First, we visualize the presence for observed sites:
ggplot(moose, aes(color = presence)) +
geom_sf(size = 2) +
scale_color_viridis_d(option = "H") +
theme_gray(base_size = 14)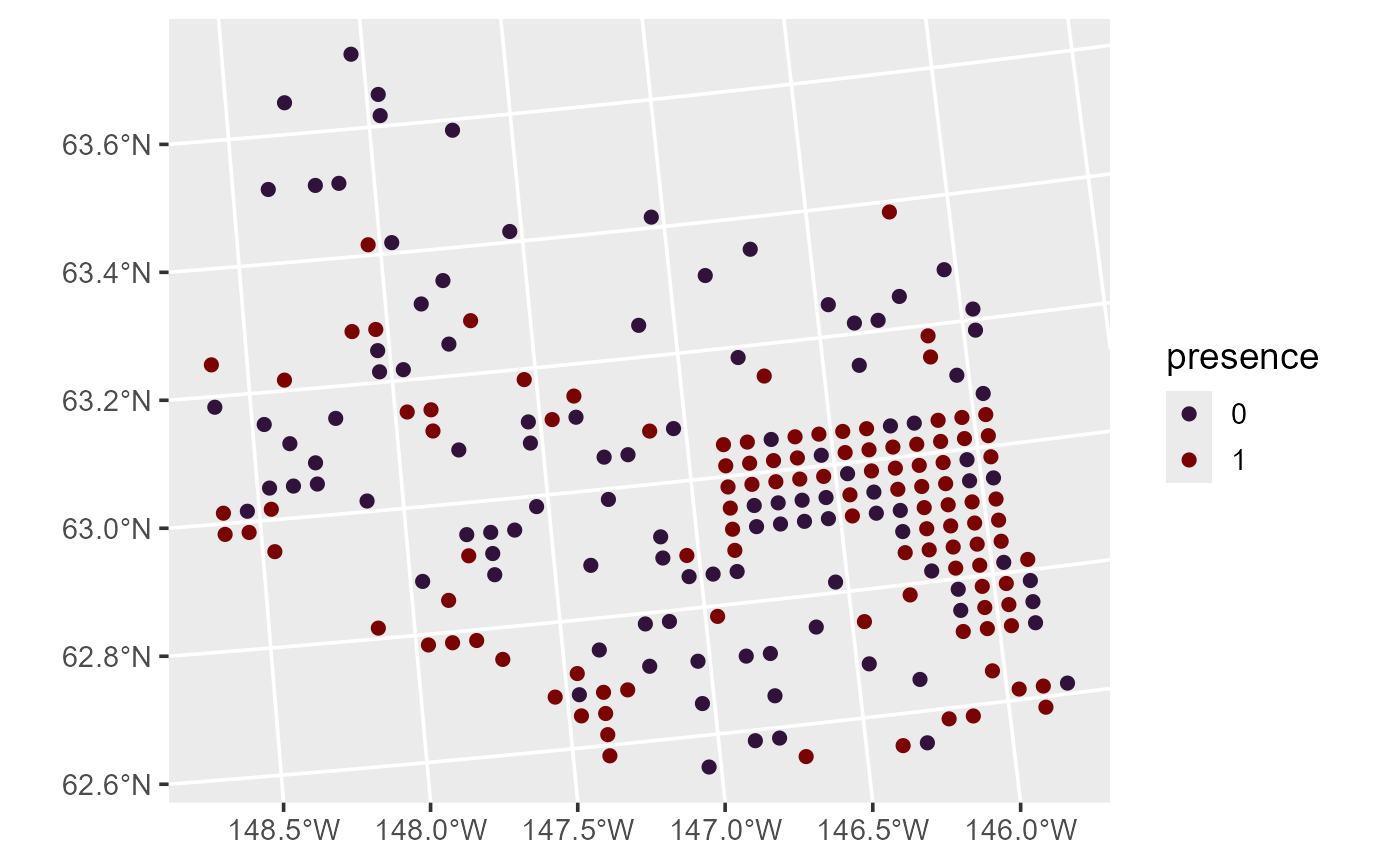
Moose presence.
There is spatial patterning in the distribution of moose presence, as moose tend to be present in the eastern and southwestern parts of the domain. This spatial pattern seems to have a directional orientation, seemingly strongest in the northwest-to-southeast direction.
Next, we visualize the elevation for each strata:
ggplot(moose, aes(color = elev)) +
geom_sf(size = 1) +
facet_grid(~ strat) +
scale_color_viridis_c() +
scale_y_continuous(breaks = seq(62.6, 63.6, length.out = 5)) +
scale_x_continuous(breaks = seq(-148.5, -146, length.out = 3)) +
theme_gray(base_size = 12)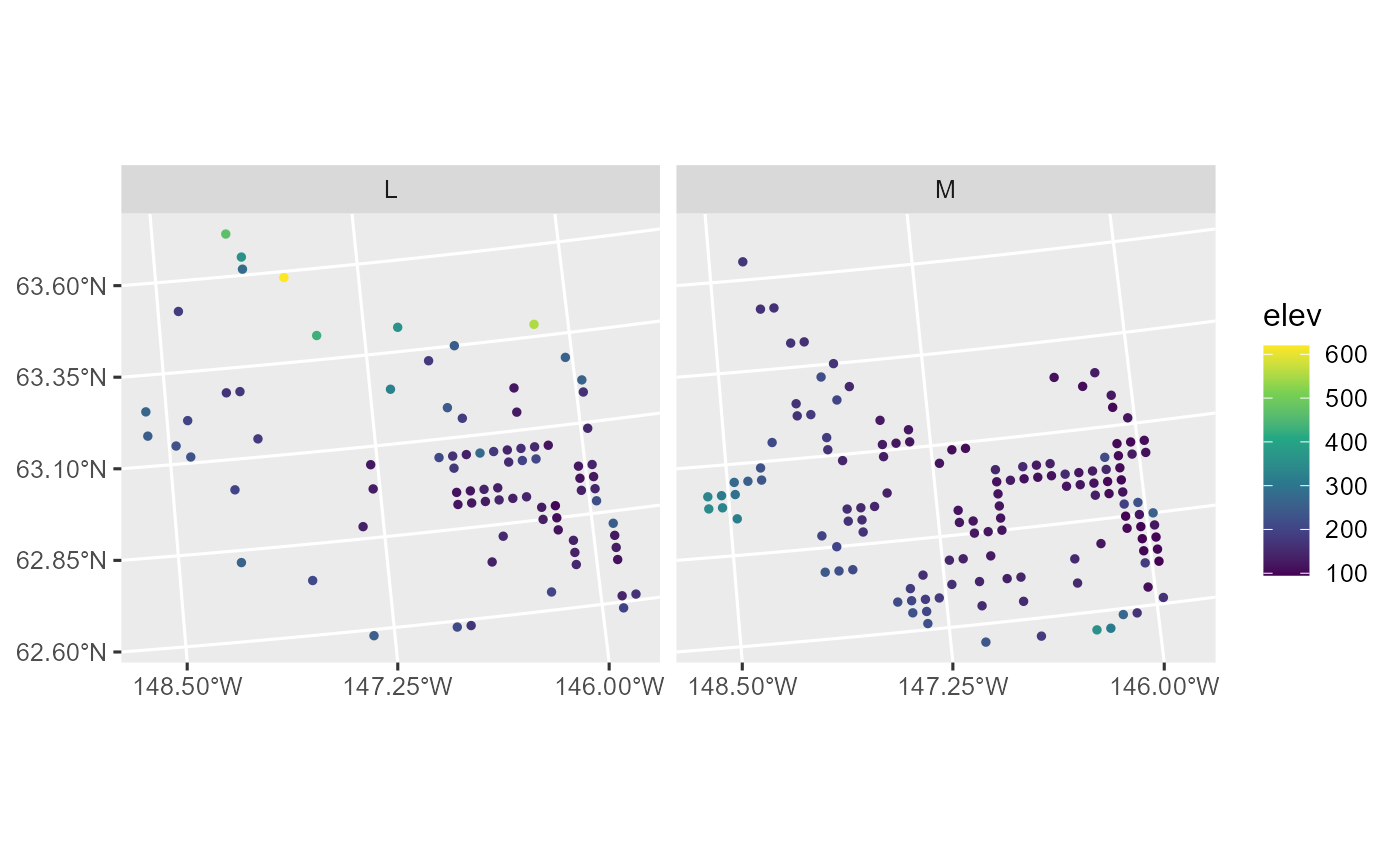
Elevation versus strata in the moose data.
There are no obvious spatial patterns in the assignment of sites to strata, while elevation is highest on the boundaries of the domain and lowest in the middle of the domain. There is spatial patterning in the distribution of moose presence, as moose tend to be present in the eastern and southwestern parts of the domain. This spatial pattern seems to have a directional orientation, seemingly strongest in the northwest-to-southeast direction.
To quantify the effects of elev, strat, and
spatial dependence on moose presence, we fit three binomial (i.e., )
logistic regression models using spglm():
# model one
bin_mod <- spglm(
formula = presence ~ elev + strat + elev:strat,
family = "binomial",
data = moose,
spcov_type = "none"
)
# model two
bin_spmod <- spglm(
formula = presence ~ elev + strat + elev:strat,
family = "binomial",
data = moose,
spcov_type = "spherical"
)
# model three
bin_spmod_anis <- spglm(
formula = presence ~ elev + strat + elev:strat,
family = "binomial",
data = moose,
spcov_type = "spherical",
anisotropy = TRUE
)All three models have the same fixed effect structure, using
elevation, strata, and their interaction to explain moose presence. The
three models vary in their covariance structure. The first model,
bin_mod, has no spatial covariance (\(\sigma^2_{de} = 0\)). The second model,
bin_spmod has a spherical spatial covariance. The third
model, bin_spmod_anis, has a spherical spatial covariance
that incorporates geometric anisotropy. Geometric anisotropy allows the
spatial covariance to vary with direction by evaluating the spatial
covariance with \(\mathbf{H}^*\)
(instead of \(\mathbf{H}\)), a matrix
of distances between transformed coordinates that are rotated and scaled
appropriately (Schabenberger and Gotway
2017). We evaluate the three models using AIC by running
AIC(bin_mod, bin_spmod, bin_spmod_anis)#> df AIC
#> bin_mod 0 717.1627
#> bin_spmod 3 686.9031
#> bin_spmod_anis 5 677.9403The spatial models (bin_spmod,
bin_spmod_anis) have a much lower AIC than the non-spatial
model (bin_mod), which suggests that the models benefit
from incorporating spatial dependence. bin_spmod_anis has a
lower AIC than bin_spmod, which suggests that the model
benefits from incorporating directionality in the spatial dependence.
Next we inspect bin_spmod_anis and later use it to predict
moose presence probability at unobserved sites.
We summarize bin_spmod_anis using
summary():
summary(bin_spmod_anis)#>
#> Call:
#> spglm(formula = presence ~ elev + strat + elev:strat, family = "binomial",
#> data = moose, spcov_type = "spherical", anisotropy = TRUE)
#>
#> Deviance Residuals:
#> Min 1Q Median 3Q Max
#> -1.7970 -0.7348 0.3291 0.6855 1.7384
#>
#> Coefficients (fixed):
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -3.112818 1.522362 -2.045 0.04088 *
#> elev 0.010990 0.004655 2.361 0.01824 *
#> stratM 3.820320 1.176127 3.248 0.00116 **
#> elev:stratM -0.014020 0.006933 -2.022 0.04316 *
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Pseudo R-squared: 0.1119
#>
#> Coefficients (spherical spatial covariance):
#> de ie range rotate scale
#> 6.580e+00 2.220e-03 1.791e+05 2.718e+00 2.154e-01
#> attr(,"class")
#> [1] "spherical"
#>
#> Coefficients (Dispersion for binomial family):
#> dispersion
#> 1The summary() output from an spglm() model
is very similar to the summary() output from a
base-R glm() model, returning the original
function call, deviance residuals, a fixed effects coefficients table, a
pseudo R-squared (which quantifies the amount of variability explained
by the fixed effects), and covariance parameter coefficients. The fixed
effects coefficients table provides some evidence that elevation and
strata are associated with with moose presence and that the effect of
elevation varies across the two strata (all \(p~\)values \(<\) 0.05).
The area under the receiver receiver operating characteristic (AUROC)
curve quantifies the effectiveness of a classifier (Robin et al. 2011). It ranges from zero to one,
and higher values indicate better model performance. The AUROC of
bin_spmod_anis is
AUROC(bin_spmod_anis)#> [1] 0.9403199The plot() function returns graphics of several model
diagnostics. Running
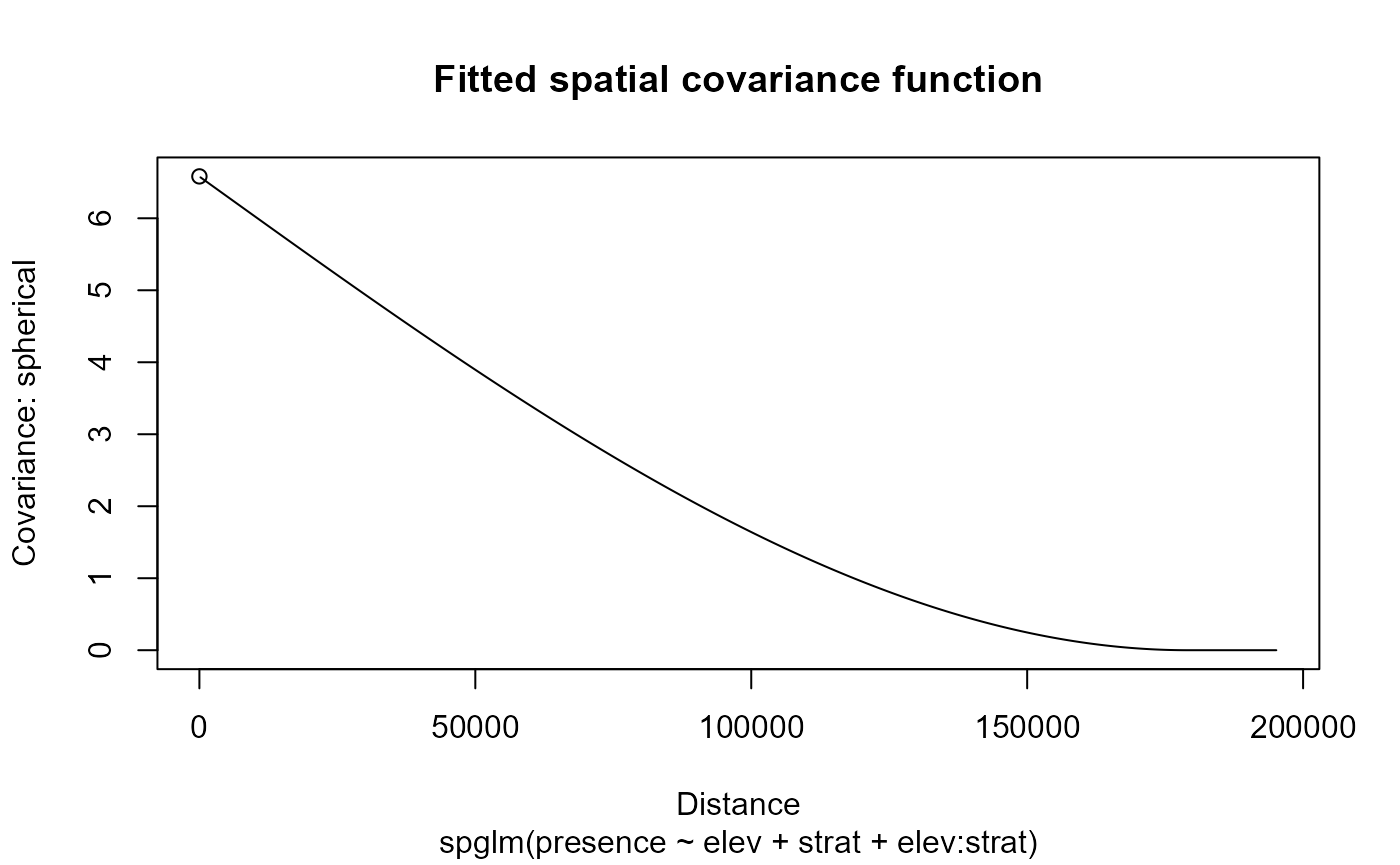
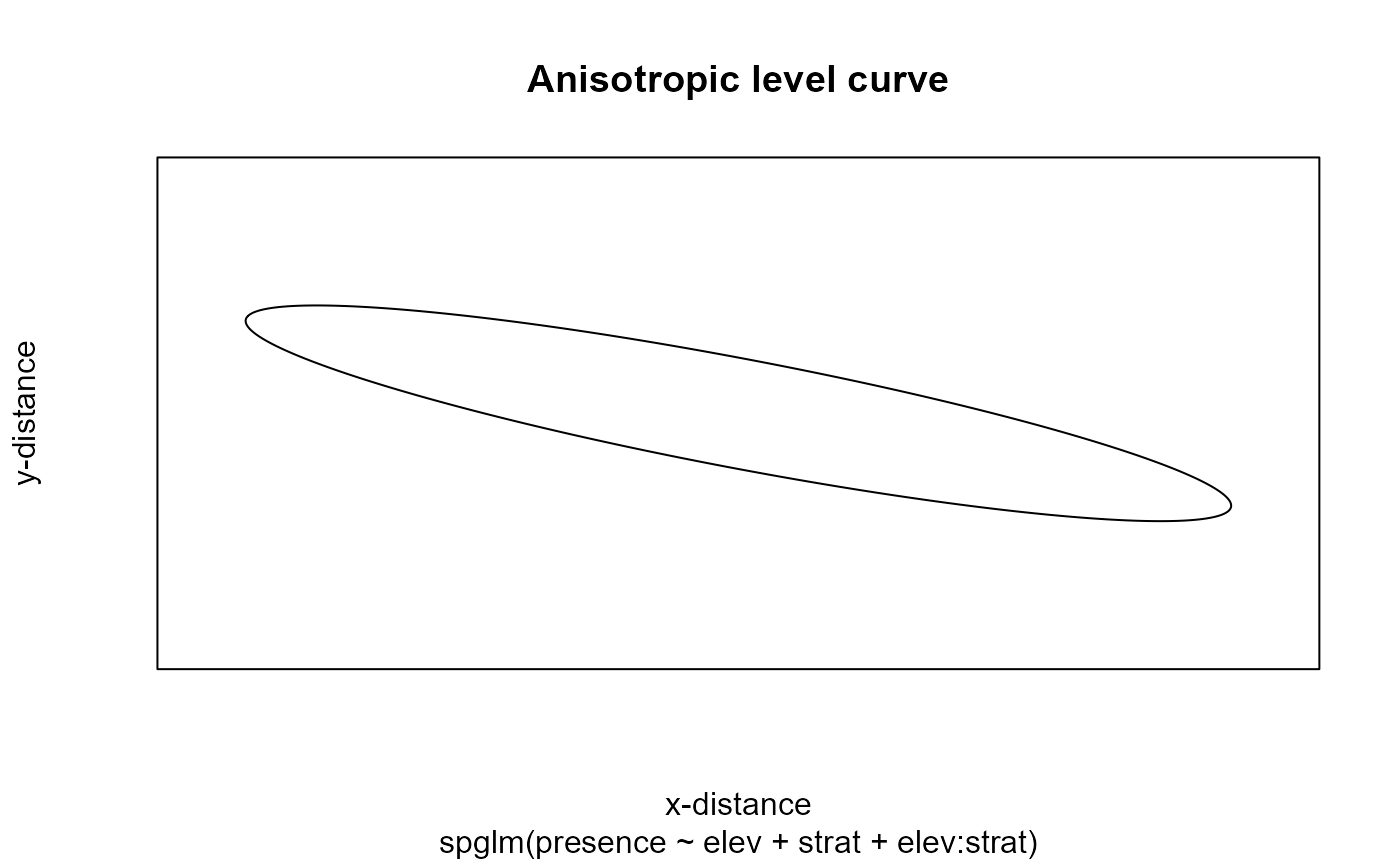
Spatial dependence is a function of distance (left) and as an anisotropic level curve of equal correlation (right).
shows that the spatial dependence is evident and appears strongest in the northwest-to-southeast direction. These findings are supported by the clear spatial patterns in the data.
Recall that the tidy(), glance(), and
augment() functions are particularly useful tools for manipulating and understanding fitted model objects. Thetidy()function tidies model output, returning atibble`
of parameter estimates (and confidence intervals):
tidy(bin_spmod_anis, conf.int = TRUE)#> # A tibble: 4 × 7
#> term estimate std.error statistic p.value conf.low conf.high
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 (Intercept) -3.11 1.52 -2.04 0.0409 -6.10 -0.129
#> 2 elev 0.0110 0.00466 2.36 0.0182 0.00187 0.0201
#> 3 elev:stratM -0.0140 0.00693 -2.02 0.0432 -0.0276 -0.000431
#> 4 stratM 3.82 1.18 3.25 0.00116 1.52 6.13By default, the fixed effects estimates are returned, but covariance
parameter estimates are returned via the effects
argument:
tidy(bin_spmod_anis, effects = "spcov")#> # A tibble: 5 × 3
#> term estimate is_known
#> <chr> <dbl> <lgl>
#> 1 de 6.58 FALSE
#> 2 ie 0.00222 FALSE
#> 3 range 179082. FALSE
#> 4 rotate 2.72 FALSE
#> 5 scale 0.215 FALSEThe is_known column indicates whether covariance
parameters were assumed known, which is possible to specify using the
spcov_initial argument to spglm(). Here, all
parameters were assumed unknown and then estimated.
The glance() function glances at model fit, returning a
tibble of model fit statistics:
glance(bin_spmod_anis)#> # A tibble: 1 × 10
#> n p npar value AIC AICc BIC logLik deviance pseudo.r.squared
#> <int> <dbl> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 218 4 5 668. 678. 678. 695. -334. 161. 0.112glances() can be used to glance at multiple models
simultaneously and sorts models by ascending AICc:
glances(bin_mod, bin_spmod, bin_spmod_anis)#> # A tibble: 3 × 11
#> model n p npar value AIC AICc BIC logLik deviance
#> <chr> <int> <dbl> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 bin_spmod_anis 218 4 5 668. 678. 678. 695. -334. 161.
#> 2 bin_spmod 218 4 3 681. 687. 687. 697. -340. 161.
#> 3 bin_mod 218 4 0 717. 717. 717. 717. -359. 294.
#> # ℹ 1 more variable: pseudo.r.squared <dbl>The augment() function augments the data with model
diagnostics:
aug_mod <- augment(bin_spmod_anis)
aug_mod#> Simple feature collection with 218 features and 8 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: 269085 ymin: 1416151 xmax: 419057.4 ymax: 1541016
#> Projected CRS: NAD83 / Alaska Albers
#> # A tibble: 218 × 9
#> presence elev strat .fitted .resid .hat .cooksd .std.resid
#> * <fct> <dbl> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 0 469. L -1.91 -0.527 0.0923 0.00777 -0.553
#> 2 0 362. L -3.58 -0.235 0.00973 0.000137 -0.236
#> 3 0 173. M -3.49 -0.246 0.00136 0.0000205 -0.246
#> 4 0 280. L -4.41 -0.155 0.00187 0.0000113 -0.155
#> 5 0 620. L -0.964 -0.804 0.360 0.142 -1.01
#> 6 0 164. M -2.64 -0.372 0.00286 0.0000996 -0.373
#> 7 0 164. M -2.24 -0.449 0.00397 0.000202 -0.450
#> 8 0 186. L -2.51 -0.396 0.00486 0.000192 -0.397
#> 9 0 362. L -2.73 -0.355 0.0211 0.000692 -0.359
#> 10 0 430. L -1.62 -0.600 0.0871 0.00941 -0.628
#> # ℹ 208 more rows
#> # ℹ 1 more variable: geometry <POINT [m]>where .fitted are fitted values (the estimated \(f^{-1}(\mathbf{w})\), or moose presence
probability), .resid are response residuals,
.hat values indicate leverage, .cooksd values
indicate Cook’s distance (i.e., influence), and .std.resid
are standardized residuals.
The moose_preds data contains survey sites that were not
sampled. We can use bin_spmod_anis to make predictions of
the underlying probabilities of moose presence at these sites using
predict() or augment(). predict()
and augment() return the same predictions but
augment() augments the prediction data with the
predictions:
data("moose_preds")
spmod_anis_preds <- predict(
bin_spmod_anis,
newdata = moose_preds,
type = "response",
interval = "prediction"
)
head(spmod_anis_preds)#> fit lwr upr
#> 1 0.53625613 0.150268897 0.8831963
#> 2 0.45236331 0.106721924 0.8509940
#> 3 0.09535359 0.010375867 0.5144805
#> 4 0.10772261 0.022873507 0.3837176
#> 5 0.83772891 0.466479245 0.9682358
#> 6 0.03276524 0.003867679 0.2281264
aug_pred <- augment(
bin_spmod_anis,
newdata = moose_preds,
type.predict = "response",
interval = "prediction"
)
aug_pred#> Simple feature collection with 100 features and 5 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: 269386.2 ymin: 1418453 xmax: 419976.2 ymax: 1541763
#> Projected CRS: NAD83 / Alaska Albers
#> # A tibble: 100 × 6
#> elev strat .fitted .lower .upper geometry
#> * <dbl> <chr> <dbl> <dbl> <dbl> <POINT [m]>
#> 1 143. L 0.536 0.150 0.883 (401239.6 1436192)
#> 2 324. L 0.452 0.107 0.851 (352640.6 1490695)
#> 3 158. L 0.0954 0.0104 0.514 (360954.9 1491590)
#> 4 221. M 0.108 0.0229 0.384 (291839.8 1466091)
#> 5 209. M 0.838 0.466 0.968 (310991.9 1441630)
#> 6 218. L 0.0328 0.00387 0.228 (304473.8 1512103)
#> 7 127. L 0.0381 0.00511 0.234 (339011.1 1459318)
#> 8 122. L 0.0747 0.00983 0.396 (342827.3 1463452)
#> 9 191 L 0.375 0.0410 0.893 (284453.8 1502837)
#> 10 105. L 0.142 0.0197 0.577 (391343.9 1483791)
#> # ℹ 90 more rowsEstimated moose presence probability for the moose data
(obtained via fitted()) and predicted moose presence
probability for the moose_preds data (obtained via
predict() or augment()) are overlain below and
share similar patterns:
aug_mod$type <- "mod"
aug_pred$type <- "pred"
keep_cols <- c(".fitted", "type")
aug_combined <- rbind(aug_mod[, keep_cols], aug_pred[, keep_cols])
ggplot(aug_combined, aes(color = .fitted, shape = type)) +
geom_sf(size = 2) +
scale_color_viridis_c(option = "H") +
theme_gray(base_size = 14)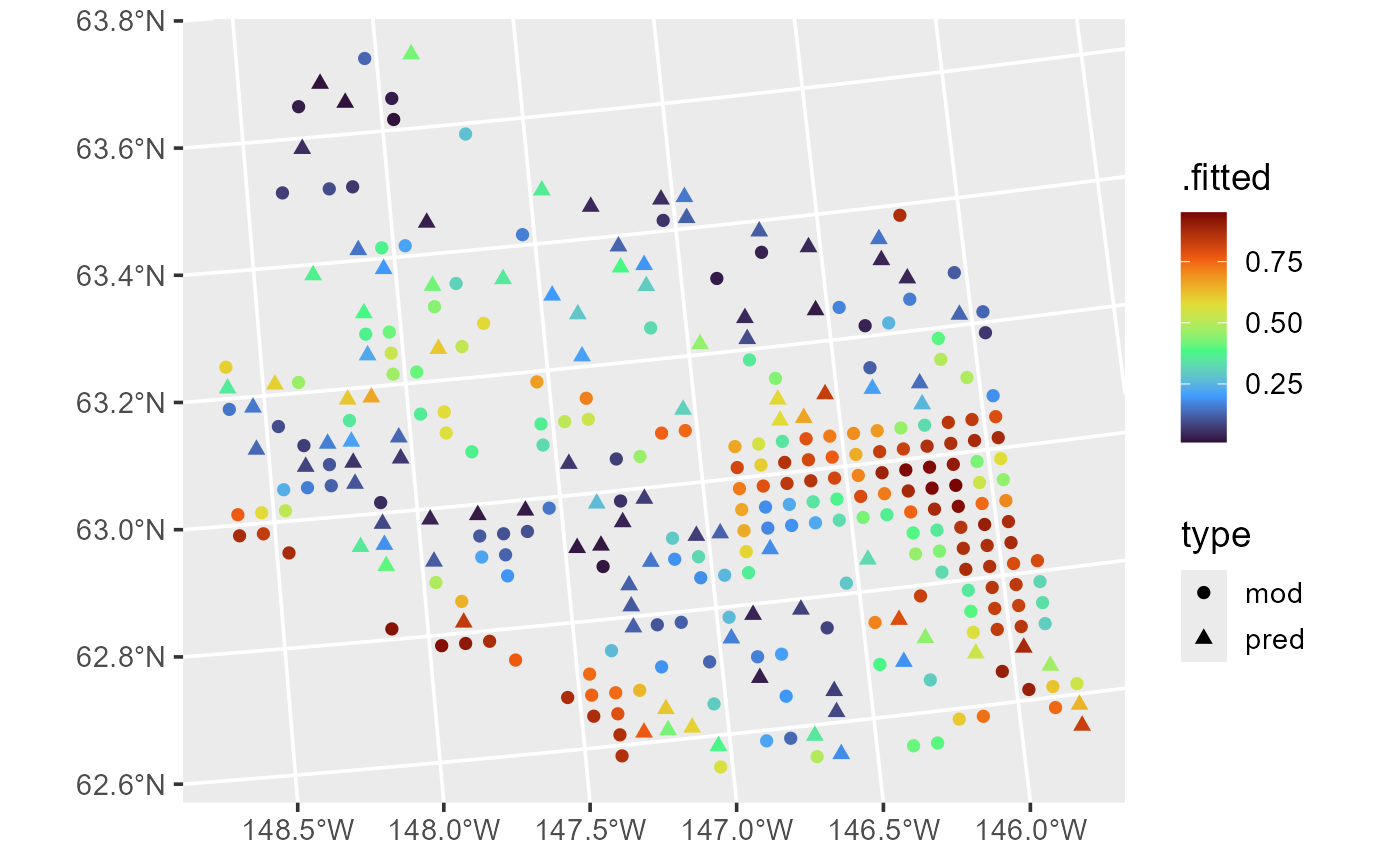
Fitted values and predictions for moose presence probability.
An application to count data
The moose data also contain the number of moose observed
at each site:
ggplot(moose, aes(color = count)) +
geom_sf(size = 2) +
scale_color_viridis_c(option = "H") +
theme_gray(base_size = 14)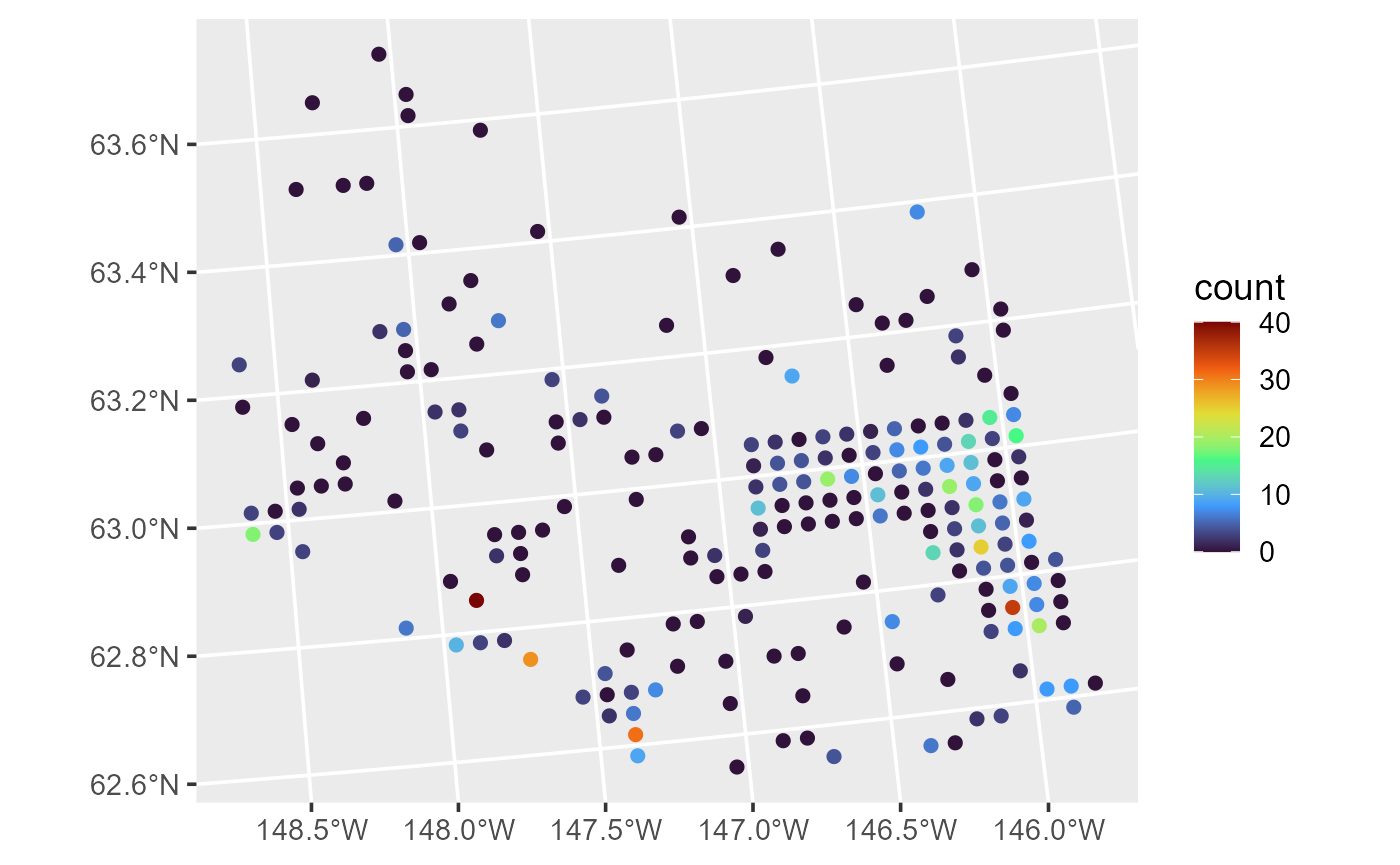
Moose counts.
The moose counts are similarly distributed as moose presence, highest
in the eastern and southwestern parts of the domain. We compare two
count models, Poisson and negative binomial. The Poisson model assumes
the underlying process generating the counts has the same mean and
variance while the negative binomial model allows for overdispersion
(i.e., the variance is greater than the mean). We are interested in
quantifying the effects of elev and strat on
moose count, albeit using a slightly different approach
than we did for moose presence. Here, we will model
elevation as a fixed effect and allow this effect to change between
strata, but we will model strata as a random effect to highlight
additional flexibility of the spmodel package. We fit
relevant Poisson and negative binomial models with a Matérn spatial
covariance using spglm():
count_mod_pois <- spglm(
count ~ elev + elev:strat,
family = "poisson",
data = moose,
spcov_type = "matern",
random = ~ (1 | strat)
)
count_mod_nb <- spglm(
count ~ elev + elev:strat,
family = "nbinomial",
data = moose,
spcov_type = "matern",
random = ~ (1 | strat)
)Random effects are specified in spmodel via the
random argument using similar syntax as the commonly used
lme4 (Bates et al. 2015) and
nlme (Pinheiro and Bates
2006) R packages for non-spatial mixed models.
In spmodel, ~ strat is short-hand for
~ (1 | strat). Random effects alter the covariance
structure of the model, building additional correlation into the model
for sites sharing a level of the random effect (here, sites within the
same strata). More formally, when incorporating a random effect, the
spatial generalized linear model becomes \[\begin{equation}\label{eq:spglm-rand}
f(\boldsymbol{\mu}) \equiv \mathbf{w} = \mathbf{X} \boldsymbol{\beta} +
\mathbf{Z}\mathbf{v} + \boldsymbol{\tau} + \boldsymbol{\epsilon},
\end{equation}\] where \(\mathbf{Z}\) is a design matrix that
indexes the random effects, \(\mathbf{v}\), and \(\text{Cov}(\mathbf{v}) =
\sigma^2_{v}\mathbf{I}\). Then the covariance matrix, \(\boldsymbol{\Sigma}\), becomes \(\sigma^2_v \mathbf{Z} \mathbf{Z}^\top +
\sigma^2_{de}\mathbf{R} + \sigma^2_{ie}\mathbf{I}\).
Previously we compared models using AIC(), but another
way to compare models is by leave-one-out cross validation (Hastie et al. 2009). In leave-one-out cross
validation, each observation is held-out and the model is re-fit and
used to predict the held-out observation. Then a loss statistic is
computed that compares each prediction to its true value. We calculate
mean-squared-prediction error (MSPE) using leave-one-out cross
validation for both the Poisson and negative binomial model by
running
loocv(count_mod_pois)#> # A tibble: 1 × 3
#> bias MSPE RMSPE
#> <dbl> <dbl> <dbl>
#> 1 1.37 32.0 5.66
loocv(count_mod_nb)#> # A tibble: 1 × 3
#> bias MSPE RMSPE
#> <dbl> <dbl> <dbl>
#> 1 0.291 27.1 5.20The negative binomial model has a lower MSPE, which suggests that incorporating overdispersion improves model fit here. Tidying this fitted model we see that there is some evidence elevation is related to moose counts and that this effect changes across strata (all \(p~\)values \(< 0.1\)):
tidy(count_mod_nb)#> # A tibble: 3 × 5
#> term estimate std.error statistic p.value
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 (Intercept) -0.301 1.50 -0.200 0.841
#> 2 elev 0.00590 0.00300 1.96 0.0494
#> 3 elev:stratM -0.00717 0.00408 -1.76 0.0789The varcomp() function in spmodel
apportions model variability into fixed and random components:
varcomp(count_mod_nb)#> # A tibble: 4 × 2
#> varcomp proportion
#> <chr> <dbl>
#> 1 Covariates (PR-sq) 0.0329
#> 2 de 0.498
#> 3 ie 0.00174
#> 4 1 | strat 0.467Most model variability is explained by the spatially dependent random
error (de) and the random effect for strata
(1 | strat). Running
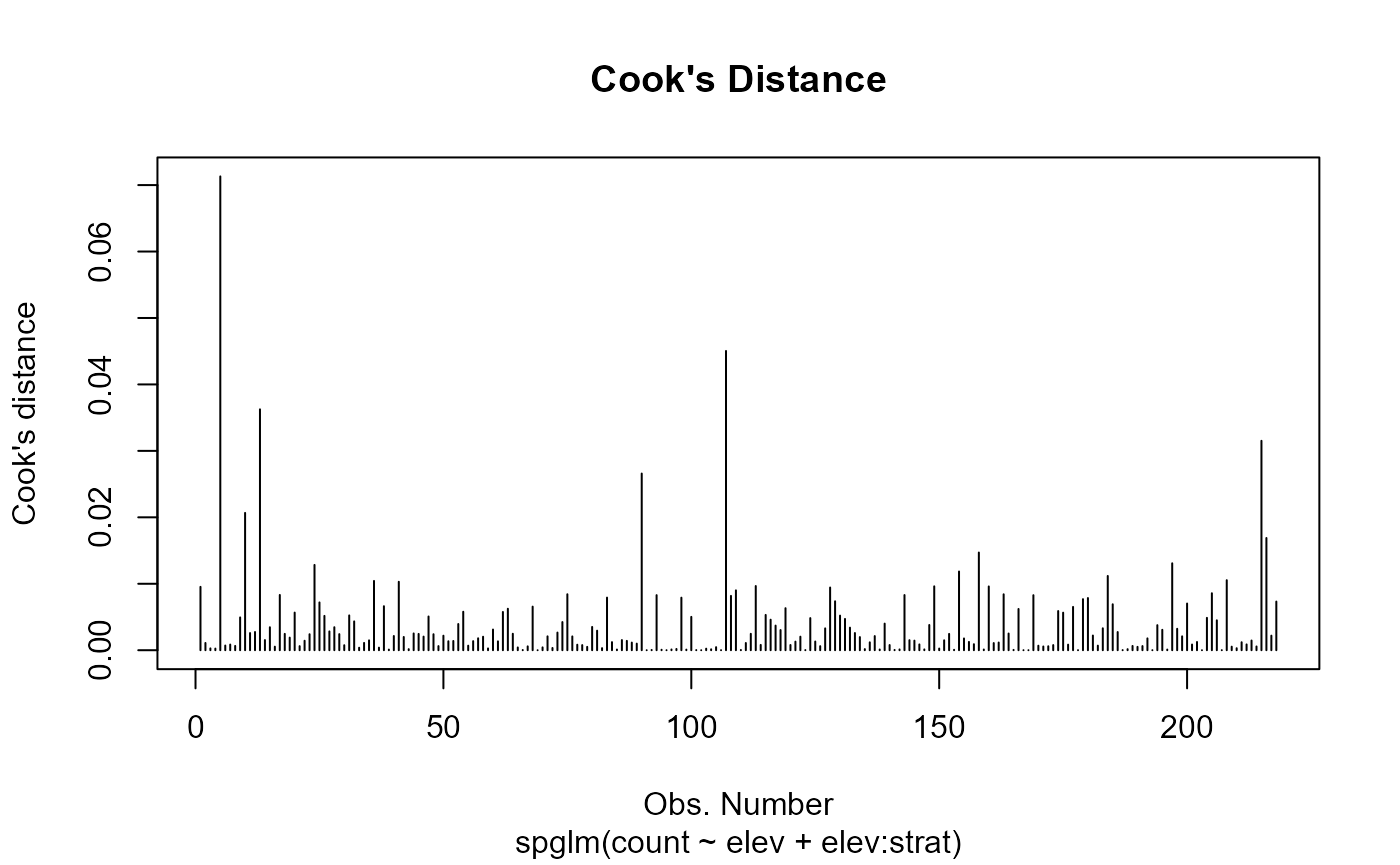

Model diagnostics.
shows observations of high influence or leverage and that the standardized residuals tend to be spread out around zero.
Estimated mean moose counts for the moose data (obtained
via fitted()) and predicted mean moose counts for the
moose_preds data (obtained via predict() or
augment()) share similar patterns and are overlain by
running
aug_mod <- augment(count_mod_nb)
aug_pred <- augment(count_mod_nb, newdata = moose_preds, type.predict = "response")
aug_mod$type <- "mod"
aug_pred$type <- "pred"
keep_cols <- c(".fitted", "type")
aug_combined <- rbind(aug_mod[, keep_cols], aug_pred[, keep_cols])
ggplot(aug_combined, aes(color = .fitted, shape = type)) +
geom_sf(size = 2) +
scale_color_viridis_c(option = "H") +
theme_gray(base_size = 18)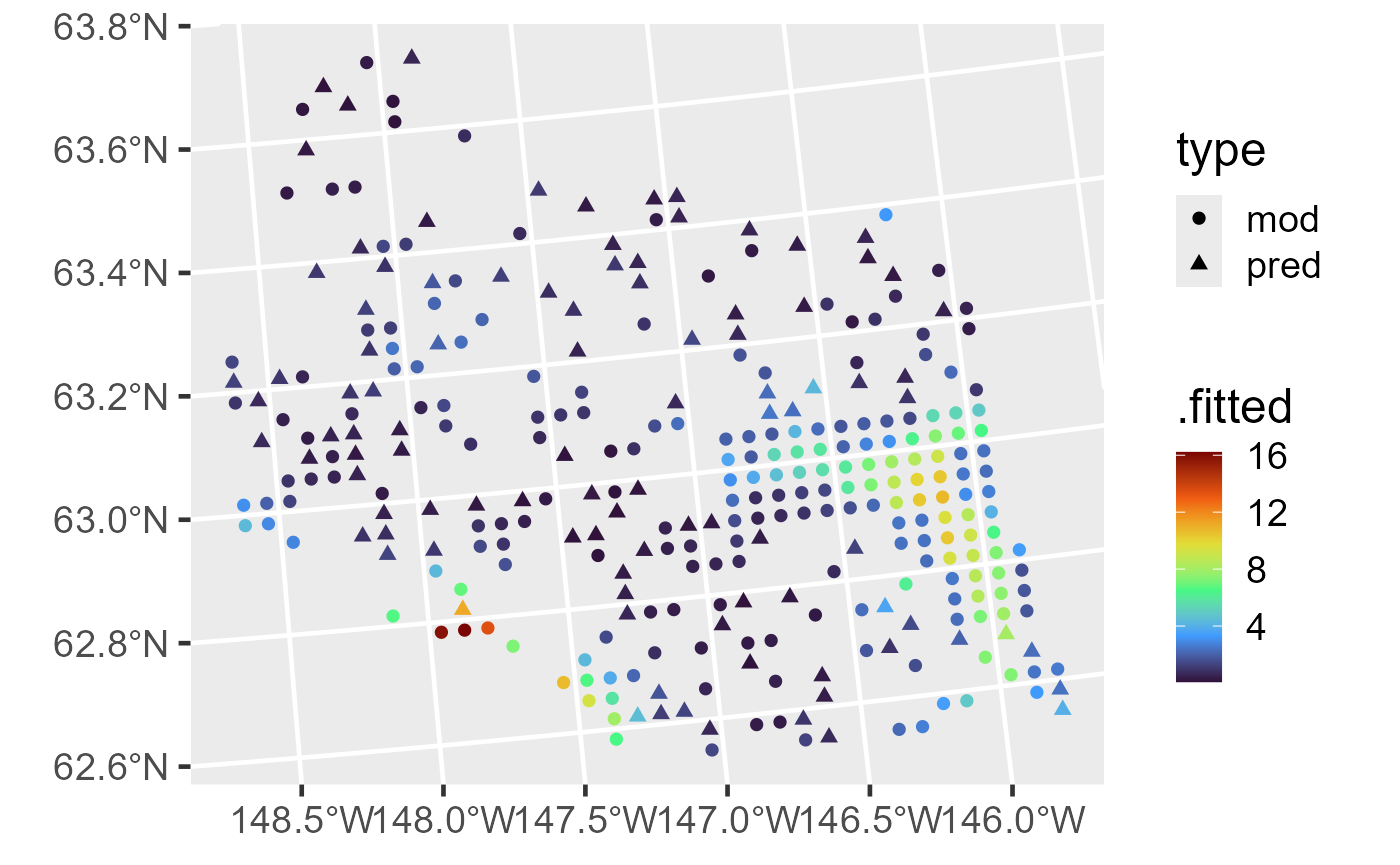
Fitted values and predictions for moose counts.
Discussion
Throughout this vignette, we have shown several features
spmodel offers, including a novel application of the
Laplace approximation, similarity to base R’s
glm() function, over a dozen spatial covariance functions,
a variety of tools available to evaluate models, inspect model
diagnostics, and make predictions using ubiquitous base
R functions (e.g., summary(),
plot(), and predict()) and more. Spatial
generalized linear models for point-referenced data (i.e., generalized
geostatistical models) are fit using the spglm() function.
Spatial generalized linear models for areal data (i.e., generalized
spatial autoregressive models) are fit using the spgautor()
function. Both functions share common structure and syntax. Spatial data
are simulated in spmodel by adding an sp
prefix to commonly used base R simulation functions
(e.g., sprbinom()).
We appreciate feedback from users regarding spmodel. To
learn more about how to provide feedback or contribute to
spmodel, please visit our GitHub repository at https://github.com/USEPA/spmodel.