A Detailed Guide to spmodel
Michael Dumelle, Matt Higham, and Jay M. Ver Hoef
Source:vignettes/articles/guide.Rmd
guide.RmdIntroduction
spmodel is an R package used to fit,
summarize, and predict for a variety of spatial statistical models. The
vignette provides an introduction to both the basic and advanced
features of the spmodel package coupled with a brief
theoretical explanation of the methods. First we give a brief
theoretical introduction to spatial linear models. Then we outline the
variety of methods used to estimate the parameters of spatial linear
models. Then we explain how to obtain predictions at unobserved
locations. Then we detail some advanced modeling features, including
random effects, partition factors, anisotropy, big data approaches, and
spatial generalized linear models. Finally we end with a short
discussion. Before proceeding, we load spmodel by
running
If using spmodel in a formal publication or report,
please cite it. Citing spmodel lets us devote more
resources to the package in the future. We view the spmodel
citation by running
citation(package = "spmodel")#> To cite spmodel in publications use:
#>
#> Dumelle M, Higham M, Ver Hoef JM (2023). spmodel: Spatial statistical
#> modeling and prediction in R. PLOS ONE 18(3): e0282524.
#> https://doi.org/10.1371/journal.pone.0282524
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Article{,
#> title = {{spmodel}: Spatial statistical modeling and prediction in {R}},
#> author = {Michael Dumelle and Matt Higham and Jay M. {Ver Hoef}},
#> journal = {PLOS ONE},
#> year = {2023},
#> volume = {18},
#> number = {3},
#> pages = {1--32},
#> doi = {10.1371/journal.pone.0282524},
#> url = {https://doi.org/10.1371/journal.pone.0282524},
#> }We will create visualizations using ggplot2 (Wickham 2016), which we load by running
ggplot2 is only installed alongside spmodel
when dependencies = TRUE in
install.packages(), so check that the package is installed
and loaded before reproducing any of these vignette’s visualizations. We
will also show code that can be used to create interactive
visualizations of spatial data with mapview (Appelhans et al. 2022). mapview
also has many backgrounds available that contextualize spatial data with
topographical information. Before running the mapview code
interactively, make sure mapview is installed and
loaded.
spmodel contains various methods for generic functions
defined outside of spmodel. To find relevant documentation
for these methods, run help("generic.spmodel", "spmodel")
(e.g., help("summary.spmodel", "spmodel"),
help("predict.spmodel", "spmodel"), etc.). Note that
?generic.spmodel is shorthand for
help("generic.spmodel", "spmodel"). We provide more details
and examples regarding these methods and generics throughout this
vignette. For a full list of spmodel functions available,
see spmodel’s documentation manual.
The Spatial Linear Model
Statistical linear models are often parameterized as \[ \mathbf{y} = \mathbf{X} \boldsymbol{\beta} + \boldsymbol{\epsilon}, \] where for a sample size \(n\), \(\mathbf{y}\) is an \(n \times 1\) column vector of response variables, \(\mathbf{X}\) is an \(n \times p\) design (model) matrix of explanatory variables, \(\boldsymbol{\beta}\) is a \(p \times 1\) column vector of fixed effects controlling the impact of \(\mathbf{X}\) on \(\mathbf{y}\), and \(\boldsymbol{\epsilon}\) is an \(n \times 1\) column vector of random errors. We typically assume that \(\text{E}(\boldsymbol{\epsilon}) = \mathbf{0}\) and \(\text{Cov}(\boldsymbol{\epsilon}) = \sigma^2_\epsilon \mathbf{I}\), where \(\text{E}(\cdot)\) denotes expectation, \(\text{Cov}(\cdot)\) denotes covariance, \(\sigma^2_\epsilon\) denotes a variance parameter, and \(\mathbf{I}\) denotes the identity matrix.
The model \(\mathbf{y} = \mathbf{X} \boldsymbol{\beta} + \boldsymbol{\epsilon}\) assumes the elements of \(\mathbf{y}\) are uncorrelated. Typically for spatial data, elements of \(\mathbf{y}\) are correlated, as observations close together in space tend to be more similar than observations far apart (Tobler 1970). Failing to properly accommodate the spatial dependence in \(\mathbf{y}\) can cause researchers to draw incorrect conclusions about their data. To accommodate spatial dependence in \(\mathbf{y}\), an \(n \times 1\) spatial random effect, \(\boldsymbol{\tau}\), is added to the linear model, yielding the model \[ \mathbf{y} = \mathbf{X} \boldsymbol{\beta} + \boldsymbol{\tau} + \boldsymbol{\epsilon}, \] where \(\boldsymbol{\tau}\) is independent of \(\boldsymbol{\epsilon}\), \(\text{E}(\boldsymbol{\tau}) = \mathbf{0}\), \(\text{Cov}(\boldsymbol{\tau}) = \sigma^2_\tau \mathbf{R}\), \(\mathbf{R}\) is a matrix that determines the spatial dependence structure in \(\mathbf{y}\) and depends on a range parameter, \(\phi\). We discuss \(\mathbf{R}\) in more detail shortly. The parameter \(\sigma^2_\tau\) is called the spatially dependent random error variance or partial sill. The parameter \(\sigma^2_\epsilon\) is called the spatially independent random error variance or nugget. These two variance parameters are henceforth more intuitively written as \(\sigma^2_{de}\) and \(\sigma^2_{ie}\), respectively. The covariance of \(\mathbf{y}\) is denoted \(\boldsymbol{\Sigma}\) and given by \(\sigma^2_{de} \mathbf{R} + \sigma^2_{ie} \mathbf{I}\). The parameters that compose this covariance are contained in the vector \(\boldsymbol{\theta}\), which is called the covariance parameter vector.
The model \(\mathbf{y} = \mathbf{X}
\boldsymbol{\beta} + \boldsymbol{\tau} + \boldsymbol{\epsilon}\)
is called the spatial linear model. The spatial linear model applies to
both point-referenced and areal (i.e., lattice) data. Spatial data are
point-referenced when the elements in \(\mathbf{y}\) are observed at
point-locations indexed by x-coordinates and y-coordinates on a
spatially continuous surface with an infinite number of locations. The
splm() function is used to fit spatial linear models for
point-referenced data (these are sometimes called geostatistical
models). One spatial covariance function available in
splm() is the exponential spatial covariance function,
which has an \(\mathbf{R}\) matrix
given by \[
\mathbf{R} = \exp(-\mathbf{M} / \phi),
\] where \(\mathbf{M}\) is a
matrix of Euclidean distances among observations. Recall that \(\phi\) is the range parameter, controlling
the behavior of of the covariance function as a function of distance.
Parameterizations for splm() spatial covariance types and
their \(\mathbf{R}\) matrices can be
seen by running help("splm", "spmodel") or
vignette("technical", "spmodel"). Some of these spatial
covariance types (e.g., Matérn) depend on an extra parameter beyond
\(\sigma^2_{de}\), \(\sigma^2_{ie}\), and \(\phi\).
Spatial data are areal when the elements in \(\mathbf{y}\) are observed as part of a
finite network of polygons whose connections are indexed by a
neighborhood structure. For example, the polygons may represent counties
in a state that are neighbors if they share at least one boundary. Areal
data are often equivalently called lattice data (Cressie 1993). The spautor()
function is used to fit spatial linear models for areal data (these are
sometimes called spatial autoregressive models). One spatial
autoregressive covariance function available in spautor()
is the simultaneous autoregressive spatial covariance function, which
has an \(\mathbf{R}\) matrix given by
\[
\mathbf{R} = [(\mathbf{I} - \phi \mathbf{W})(\mathbf{I} - \phi
\mathbf{W})^\top]^{-1},
\] where \(\mathbf{W}\) is a
weight matrix describing the neighborhood structure in \(\mathbf{y}\). Parameterizations for
spautor() spatial covariance types and their \(\mathbf{R}\) matrices can be seen by
running help("spautor", "spmodel") or
vignette("technical", "spmodel").
One way to define \(\mathbf{W}\) is through queen contiguity (Anselin, Syabri, and Kho 2010). Two observations are queen contiguous if they share a boundary. The \(ij\)th element of \(\mathbf{W}\) is then one if observation \(i\) and observation \(j\) are queen contiguous and zero otherwise. Observations are not considered neighbors with themselves, so each diagonal element of \(\mathbf{W}\) is zero.
Sometimes each element in the weight matrix \(\mathbf{W}\) is divided by its respective row sum. This is called row-standardization. Row-standardizing \(\mathbf{W}\) has several benefits, which are discussed in detail by Ver Hoef et al. (2018).
Model Fitting
In this section, we show how to use the splm() and
spautor() functions to estimate parameters of the spatial
linear model. We also explore diagnostic tools in spmodel
that evaluate model fit. The splm() and
spautor() functions share similar syntactic structure with
the lm() function used to fit linear models without spatial
dependence. splm() and spautor() generally
require at least three arguments:
-
formula: a formula that describes the relationship between the response variable (\(\mathbf{y}\)) and explanatory variables (\(\mathbf{X}\)) -
formulainsplm()is the same asformulainlm() -
data: adata.frameorsfobject that contains the response variable, explanatory variables, and spatial information -
spcov_type: the spatial covariance type ("exponential","matern","car", etc)
If data is an sf (Pebesma 2018) object, spatial information is
stored in the object’s geometry. If data is a
data.frame, then the x-coordinates and y-coordinates must
be provided via the xcoord and ycoord
arguments (for point-referenced data) or the weight matrix must be
provided via the W argument (for areal data). The appendix
uses the caribou data, a tibble (a special
data.frame), to show how to provide spatial information via
xcoord and ycoord (in splm()) or
W (in spautor()).
In the following subsections, we use the point-referenced
moss data, an sf object that contains data on
heavy metals in mosses near a mining road in Alaska. We view the first
few rows of moss by running
moss#> Simple feature collection with 365 features and 7 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: -445884.1 ymin: 1929616 xmax: -383656.8 ymax: 2061414
#> Projected CRS: NAD83 / Alaska Albers
#> # A tibble: 365 × 8
#> sample field_dup lab_rep year sideroad log_dist2road log_Zn
#> <fct> <fct> <fct> <fct> <fct> <dbl> <dbl>
#> 1 001PR 1 1 2001 N 2.68 7.33
#> 2 001PR 1 2 2001 N 2.68 7.38
#> 3 002PR 1 1 2001 N 2.54 7.58
#> 4 003PR 1 1 2001 N 2.97 7.63
#> 5 004PR 1 1 2001 N 2.72 7.26
#> 6 005PR 1 1 2001 N 2.76 7.65
#> 7 006PR 1 1 2001 S 2.30 7.59
#> 8 007PR 1 1 2001 N 2.78 7.16
#> 9 008PR 1 1 2001 N 2.93 7.19
#> 10 009PR 1 1 2001 N 2.79 8.07
#> # ℹ 355 more rows
#> # ℹ 1 more variable: geometry <POINT [m]>We can learn more about moss by running
help("moss", "spmodel"), and we can visualize the
distribution of log zinc concentration in moss by
running
ggplot(moss, aes(color = log_Zn)) +
geom_sf() +
scale_color_viridis_c() +
theme_gray(base_size = 14)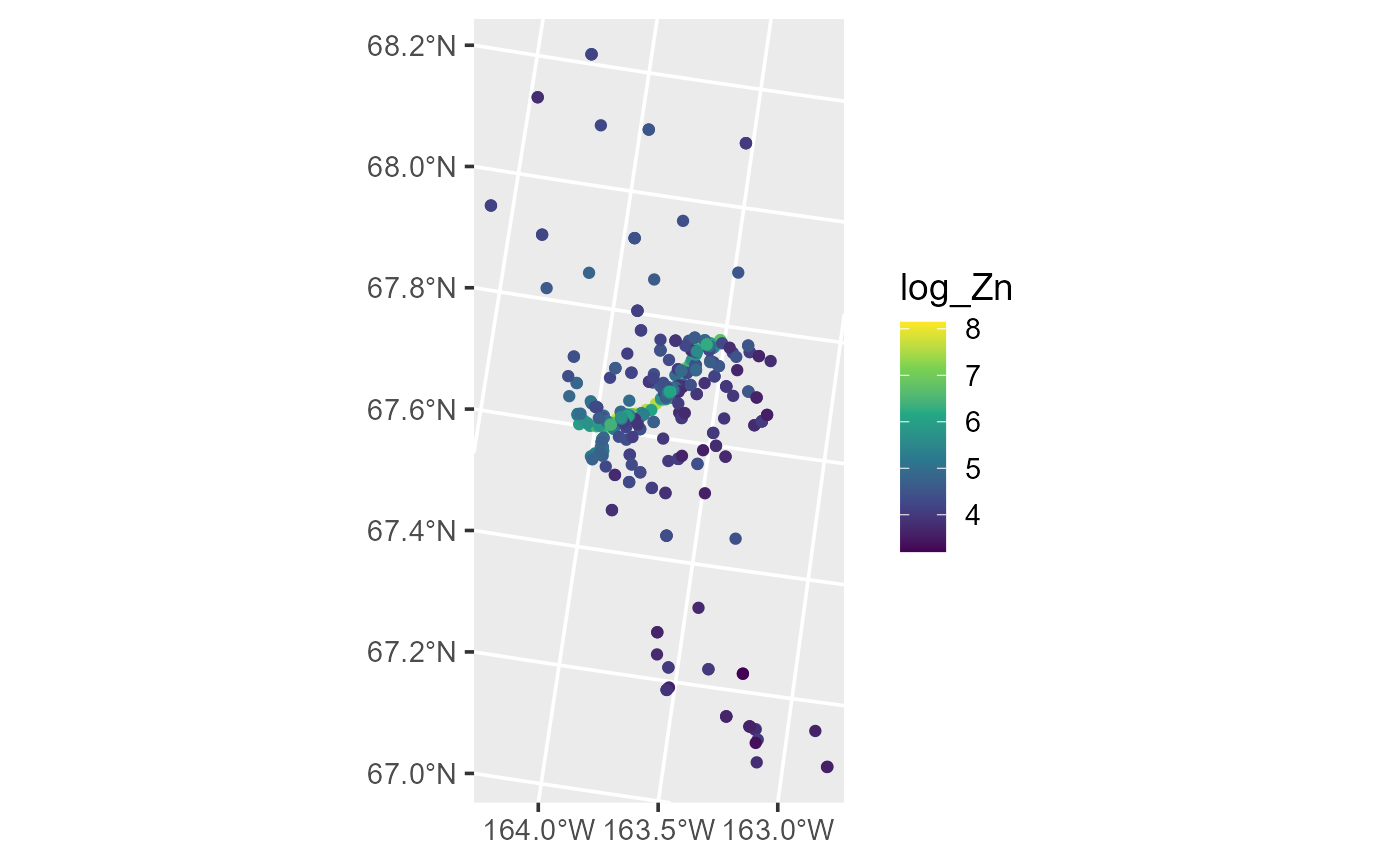
Distribution of log zinc concentration in the moss data.
Log zinc concentration can be viewed interactively in
mapview by running
mapview(moss, zcol = "log_Zn")Estimation
Generally the covariance parameters (\(\boldsymbol{\theta}\)) and fixed effects
(\(\boldsymbol{\beta}\)) of the spatial
linear model require estimation. The default estimation method in
spmodel is restricted maximum likelihood (Patterson and Thompson 1971; Harville 1977; Wolfinger,
Tobias, and Sall 1994), but the estimation method can be changed
with the estmethod argument to splm() or
spautor(). Maximum likelihood estimation is also available.
For point-referenced data, semivariogram weighted least squares (Cressie 1985) and semivariogram composite
likelihood (Curriero and Lele 1999) are
additional estimation methods. The estimation method is chosen using the
estmethod argument.
We estimate parameters of a spatial linear model regressing log zinc
concentration (log_Zn) on log distance to a haul road
(log_dist2road) using an exponential spatial covariance
function by running
spmod <- splm(log_Zn ~ log_dist2road, moss, spcov_type = "exponential")We summarize the model fit by running
summary(spmod)#>
#> Call:
#> splm(formula = log_Zn ~ log_dist2road, data = moss, spcov_type = "exponential")
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -2.6801 -1.3606 -0.8103 -0.2485 1.1298
#>
#> Coefficients (fixed):
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 9.76825 0.25216 38.74 <2e-16 ***
#> log_dist2road -0.56287 0.02013 -27.96 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Pseudo R-squared: 0.683
#>
#> Coefficients (exponential spatial covariance):
#> de ie range
#> 3.595e-01 7.897e-02 8.237e+03The fixed effects coefficient table contains estimates, standard errors, z-statistics, and asymptotic p-values for each fixed effect. From this table, we notice there is evidence that mean log zinc concentration significantly decreases with distance from the haul road (p-value < 2e-16). We see the fixed effect estimates by running
coef(spmod)#> (Intercept) log_dist2road
#> 9.7682525 -0.5628713The model summary also contains the exponential spatial covariance parameter estimates, which we can view by running
coef(spmod, type = "spcov")#> de ie range rotate scale
#> 3.595316e-01 7.896824e-02 8.236712e+03 0.000000e+00 1.000000e+00
#> attr(,"class")
#> [1] "exponential"The dependent random error variance (\(\sigma^2_{de}\)) is estimated to be
approximately 0.36 and the independent random error variance (\(\sigma^2_{ie}\)) is estimated to be
approximately 0.079. The range (\(\phi\)) is estimated to be approximately
8,237. The effective range is the distance at which the spatial
covariance is approximately zero. For the exponential covariance, the
effective range is \(3\phi\). This
means that observations whose distance is greater than 24,711 meters are
approximately uncorrelated. The rotate and
scale parameters affect the modeling of anisotropy. By
default, they are assumed to be zero and one, respectively, which means
that anisotropy is not modeled (i.e., the spatial covariance is assumed
isotropic, or independent of direction). We plot the fitted spatial
covariance function by running
plot(spmod, which = 7)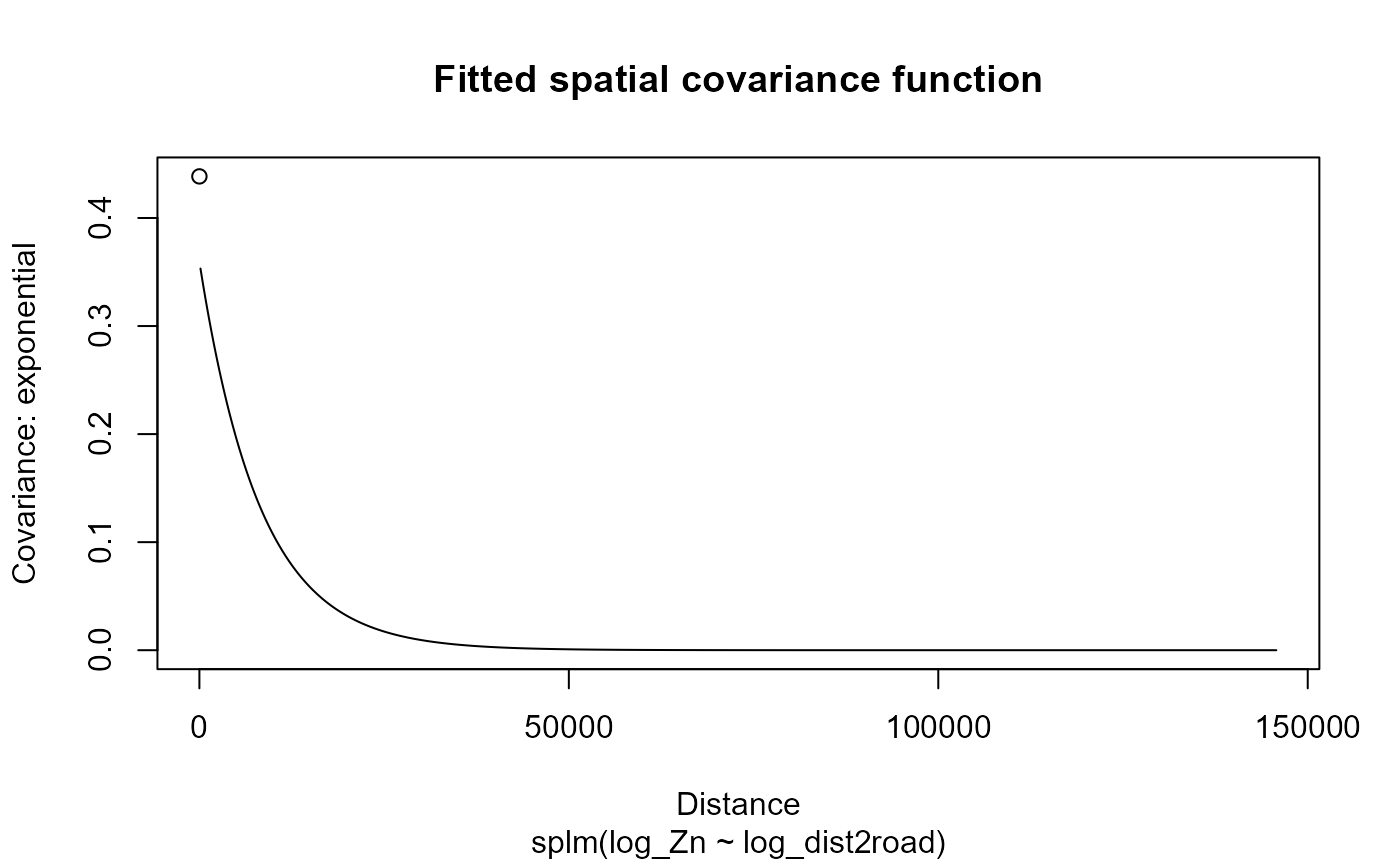
Empirical spatial covariance of fitted model.
We can learn more about the plots available for fitted models by
running help("plot.splm", "spmodel").
Model-Fit Statistics
The quality of model fit can be assessed using a variety of
statistics readily available in spmodel. The first
model-fit statistic we consider is the pseudo R-squared. The pseudo
R-squared is a generalization of the classical R-squared from
non-spatial linear models that quantifies the proportion of variability
in the data explained by the fixed effects. The pseudo R-squared is
defined as \[
PR2 = 1 -
\frac{\mathcal{D}(\boldsymbol{\hat{\Theta}})}{\mathcal{D}(\boldsymbol{\hat{\Theta}}_0)},
\] where \(\mathcal{D}(\boldsymbol{\hat{\Theta}})\) is
the deviance of the fitted model indexed by parameter vector \(\boldsymbol{\hat{\Theta}}\) and \(\mathcal{D}(\boldsymbol{\hat{\Theta}}_0)\)
is the deviance of an intercept-only model indexed by parameter vector
\(\boldsymbol{\hat{\Theta}}_0\). For
maximum likelihood, \(\hat{\boldsymbol{\Theta}} =
\{\hat{\boldsymbol{\theta}}, \hat{\boldsymbol{\beta}}\}\). For
restricted maximum likelihood \(\hat{\boldsymbol{\Theta}} =
\{\hat{\boldsymbol{\theta}}\}\).
We compute the pseudo R-squared by running We compute the pseudo R-squared by running
pseudoR2(spmod)#> [1] 0.6829687Roughly 68% of the variability in log zinc is explained by log
distance from the road. The pseudo R-squared can be adjusted to account
for the number of explanatory variables using the adjust
argument. Pseudo R-squared (and the adjusted version) is most helpful
for comparing models that have the same covariance structure.
The next two model-fit statistics we consider are the AIC and AICc. The AIC and AICc evaluate the fit of a model with a penalty for the number of parameters estimated. This penalty balances model fit and model parsimony. Lower AIC and AICc indicate a better balance of model fit and parsimony. The AICc is a correction to AIC for small sample sizes. As the sample size increases, AIC and AICc converge.
The spatial AIC and AICc are given by \[ \begin{split} \text{AIC} & = -2\ell(\hat{\boldsymbol{\Theta}}) + 2(|\hat{\boldsymbol{\Theta}}|) \\ \text{AICc} & = -2\ell(\hat{\boldsymbol{\Theta}}) + 2n(|\hat{\boldsymbol{\Theta}}|) / (n - |\hat{\boldsymbol{\Theta}}| - 1), \end{split} \] where \(\ell(\hat{\boldsymbol{\Theta}})\) is the log-likelihood of the data evaluated at the estimated parameter vector \(\hat{\boldsymbol{\Theta}}\) that maximized \(\ell(\boldsymbol{\Theta})\), \(|\hat{\boldsymbol{\Theta}}|\) is the dimension of \(\hat{\boldsymbol{\Theta}}\), and \(n\) is the sample size. As with the deviance, for maximum likelihood, \(\hat{\boldsymbol{\Theta}} = \{\hat{\boldsymbol{\theta}}, \hat{\boldsymbol{\beta}}\}\), and for restricted maximum likelihood \(\hat{\boldsymbol{\Theta}} = \{\hat{\boldsymbol{\theta}}\}\). There are some nuances to consider when comparing AIC across models: AIC comparisons between a model fit using restricted maximum likelihood and a model fit using maximum likelihood are meaningless, as the models are fit with different likelihoods; AIC comparisons between models fit using restricted maximum likelihood are only valid when the models have the same fixed effect structure; AIC comparisons between models fit using maximum likelihood are valid even when the models have different fixed effect structures (Pinheiro and Bates 2006).
Suppose we want to quantify the difference in model quality between
the spatial model and a non-spatial model using the AIC and AICc
criteria. We fit a non-spatial model in spmodel by
running
lmod <- splm(log_Zn ~ log_dist2road, moss, spcov_type = "none")This model is equivalent to one fit using lm(). We
compute the spatial AIC and AICc of the spatial model and non-spatial
model by running
AIC(spmod, lmod)#> df AIC
#> spmod 3 373.2089
#> lmod 1 633.6418
AICc(spmod, lmod)#> df AICc
#> spmod 3 373.2754
#> lmod 1 633.6528The noticeably lower AIC and AICc of the spatial model indicate that it is a better fit to the data than the non-spatial model. Recall that these AIC and AICc comparisons are valid because both models are fit using restricted maximum likelihood (the default). Similarly, one could use the BIC (Schwarz 1978) as a model comparison statistic based on the likelihood.
Another approach to comparing the fitted models is to perform
leave-one-out cross validation (Hastie et al.
2009). In leave-one-out cross validation, a single observation is
removed from the data, the model is re-fit, and a prediction is made for
the held-out observation. Then, a loss metric like
mean-squared-prediction error is computed and used to evaluate model
fit. The lower the mean-squared-prediction error, the better the model
fit. For computational efficiency, leave-one-out cross validation in
spmodel is performed by first estimating \(\boldsymbol{\theta}\) using all the data
and then re-estimating only \(\boldsymbol{\beta}\) as we predict each
removed observation. We perform leave-one-out cross validation for the
spatial and non-spatial model by running
loocv(spmod)#> # A tibble: 1 × 4
#> bias MSPE RMSPE cor2
#> <dbl> <dbl> <dbl> <dbl>
#> 1 0.00655 0.111 0.333 0.886
loocv(lmod)#> # A tibble: 1 × 4
#> bias MSPE RMSPE cor2
#> <dbl> <dbl> <dbl> <dbl>
#> 1 0.000644 0.324 0.569 0.667The noticeably lower mean-squared-prediction error of the spatial model indicates that it is a better fit to the data than the non-spatial model.
Diagnostics
In addition to model fit metrics, spmodel provides
functions that compute diagnostic metrics that help assess model
assumptions and identify unusual observations.
An observation is said to have high leverage if its combination of explanatory variable values is far from the mean vector of the explanatory variables. For a non-spatial model, the leverage of the \(i\)th observation is the \(i\)th diagonal element of the hat matrix given by \[ \mathbf{H} = \mathbf{X}(\mathbf{X}^\top\mathbf{X})^{-1}\mathbf{X}^\top . \]
For a spatial model, the leverage of the \(i\)th observation is the \(i\)th diagonal element of the spatial hat matrix given by \[ \mathbf{H}^* = (\mathbf{X}^* (\mathbf{X}^{* \top} \mathbf{X})^{-1} \mathbf{X}^{* \top}) , \] where \(\mathbf{X}^* = \boldsymbol{\Sigma}^{-1/2}\mathbf{X}\) and \(\boldsymbol{\Sigma}^{-1/2}\) is the inverse square root of the covariance matrix, \(\boldsymbol{\Sigma}\) (Montgomery, Peck, and Vining 2021). The spatial hat matrix can be viewed as the non-spatial hat matrix applied to \(\mathbf{X}^*\) instead of \(\mathbf{X}\). We compute the hat values (leverage) by running
hatvalues(spmod)Larger hat values indicate more leverage, and observations with large hat values may be unusual and warrant further investigation.
The fitted value of an observation is the estimated mean response given the observation’s explanatory variable values and the model fit: \[ \hat{\mathbf{y}} = \mathbf{X} \hat{\boldsymbol{\beta}}. \] We compute the fitted values by running
fitted(spmod)Fitted values for the spatially dependent random errors (\(\boldsymbol{\tau}\)), spatially independent
random errors (\(\boldsymbol{\epsilon}\)), and random
effects can also be obtained via fitted() by changing the
type argument.
The residuals measure each response’s deviation from its fitted value. The response residuals are given by \[ \mathbf{e}_{r} = \mathbf{y} - \hat{\mathbf{y}}. \] We compute the response residuals of the spatial model by running
residuals(spmod)The response residuals are typically not directly checked for linear model assumptions, as they have covariance closely resembling the covariance of \(\mathbf{y}\). Pre-multiplying the residuals by \(\boldsymbol{\Sigma}^{-1/2}\) yields the Pearson residuals (Myers et al. 2012): \[ \mathbf{e}_{p} = \boldsymbol{\Sigma}^{-1/2}\mathbf{e}_{r}. \] When the model is correct, the Pearson residuals have mean zero, variance approximately one, and are uncorrelated. We compute the Pearson residuals of the spatial model by running
residuals(spmod, type = "pearson")The covariance of \(\mathbf{e}_{p}\) is \((\mathbf{I} - \mathbf{H}^*)\), which is approximately \(\mathbf{I}\) for large sample sizes. Explicitly dividing \(\mathbf{e}_{p}\) by the respective diagonal element of \((\mathbf{I} - \mathbf{H}^*)\) yields the standardized residuals (Myers et al. 2012): \[ \mathbf{e}_{s} = \mathbf{e}_{p} \odot \frac{1}{\sqrt{(1 - \text{diag}(\mathbf{H}^*))}}, \] where \(\text{diag}(\mathbf{H}^*)\) denotes the diagonal of \(\mathbf{H}^*\) and \(\odot\) denotes the Hadmard (element-wise) product. We compute the standardized residuals of the spatial model by running
residuals(spmod, type = "standardized")or
rstandard(spmod)When the model is correct, the standardized residuals have mean zero, variance one, and are uncorrelated. It is common to check linear model assumptions through visualizations. We can plot the standardized residuals vs fitted values by running
plot(spmod, which = 1) # figure omittedWhen the model is correct, the standardized residuals should be evenly spread around zero with no discernible pattern. We can plot a normal QQ-plot of the standardized residuals by running
plot(spmod, which = 2) # figure omittedWhen the standardized residuals are normally distributed, they should closely follow the normal QQ-line.
An observation is said to be influential if its omission has a large impact on model fit. Typically, this is measured using Cook’s distance (Cook and Weisberg 1982). For the non-spatial model, the Cook’s distance of the \(i\)th observation is denoted \(\mathbf{D}\) and given by \[ \mathbf{D} = \frac{\mathbf{e}_{s}^2}{p} \odot \text{diag}(\mathbf{H}) \odot \frac{1}{(1 - \text{diag}(\mathbf{H}))}, \] where \(p\) is the dimension of \(\boldsymbol{\beta}\) (the number of fixed effects).
For a spatial model, the Cook’s distance of the \(i\)th observation is denoted \(\mathbf{D}^*\) and given by \[ \mathbf{D}^* = \frac{\mathbf{e}_{s}^2}{p} \odot \text{diag}(\mathbf{H}^*) \odot \frac{1}{(1 - \text{diag}(\mathbf{H}^*))} . \] A larger Cook’s distance indicates more influence, and observations with large Cook’s distance values may be unusual and warrant further investigation. We compute Cook’s distance by running
cooks.distance(spmod)The Cook’s distance versus leverage (hat values) can be visualized by running
plot(spmod, which = 6) # figure omittedThough we described the model diagnostics in this subsection using \(\boldsymbol{\Sigma}\), generally the covariance parameters are estimated and \(\boldsymbol{\Sigma}\) is replaced with \(\boldsymbol{\hat{\Sigma}}\).
The broom functions: tidy(), glance(), and
augment()
The tidy(), glance(), and
augment() functions from the broom R
package (Robinson, Hayes, and Couch 2021)
provide convenient output for many of the model fit and diagnostic
metrics discussed in the previous two sections. The tidy()
function returns a tidy tibble of the coefficient table from
summary():
tidy(spmod)#> # A tibble: 2 × 5
#> term estimate std.error statistic p.value
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 (Intercept) 9.77 0.252 38.7 0
#> 2 log_dist2road -0.563 0.0201 -28.0 0This tibble format makes it easy to pull out the coefficient names,
estimates, standard errors, z-statistics, and p-values from the
summary() output.
The glance() function returns a tidy tibble of model-fit
statistics:
glance(spmod)#> # A tibble: 1 × 10
#> n p npar value AIC AICc BIC logLik deviance pseudo.r.squared
#> <int> <dbl> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 365 2 3 367. 373. 373. 385. -184. 363. 0.683The glances() function is an extension of
glance() that can be used to look at many models
simultaneously:
glances(spmod, lmod)#> # A tibble: 2 × 11
#> model n p npar value AIC AICc BIC logLik deviance
#> <chr> <int> <dbl> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 spmod 365 2 3 367. 373. 373. 385. -184. 363.
#> 2 lmod 365 2 1 632. 634. 634. 638. -316. 363.
#> # ℹ 1 more variable: pseudo.r.squared <dbl>Finally, the augment() function augments the original
data with model diagnostics:
augment(spmod)#> Simple feature collection with 365 features and 7 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: -445884.1 ymin: 1929616 xmax: -383656.8 ymax: 2061414
#> Projected CRS: NAD83 / Alaska Albers
#> # A tibble: 365 × 8
#> log_Zn log_dist2road .fitted .resid .hat .cooksd .std.resid
#> * <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 7.33 2.68 8.26 -0.928 0.0200 0.0142 1.18
#> 2 7.38 2.68 8.26 -0.880 0.0200 0.0186 1.35
#> 3 7.58 2.54 8.34 -0.755 0.0225 0.00482 0.647
#> 4 7.63 2.97 8.09 -0.464 0.0197 0.0305 1.74
#> 5 7.26 2.72 8.24 -0.977 0.0215 0.131 3.45
#> 6 7.65 2.76 8.21 -0.568 0.0284 0.0521 1.89
#> 7 7.59 2.30 8.47 -0.886 0.0300 0.0591 1.96
#> 8 7.16 2.78 8.20 -1.05 0.0335 0.00334 0.439
#> 9 7.19 2.93 8.12 -0.926 0.0378 0.0309 1.26
#> 10 8.07 2.79 8.20 -0.123 0.0314 0.00847 0.723
#> # ℹ 355 more rows
#> # ℹ 1 more variable: geometry <POINT [m]>By default, only the columns of data used to fit the
model are returned alongside the diagnostics. All original columns of
data are returned by setting drop to
FALSE. augment() is especially powerful when
the data are an sf object because model diagnostics can be
easily visualized spatially. For example, we could subset the augmented
object so that it only includes observations whose standardized
residuals have absolute values greater than some cutoff and then
visualize them spatially. To learn more about the broom functions for
spatial linear models, run help("tidy.splm", "spmodel"),
help("glance.splm", "spmodel"), and
help("augment.splm", "spmodel").
An Areal Data Example
Next we use the seal data, an sf object
that contains the log of the estimated harbor-seal trends from abundance
data across polygons in Alaska, to provide an example of fitting a
spatial linear model for areal data using spautor(). We
view the first few rows of seal by running
seal#> Simple feature collection with 149 features and 2 fields
#> Geometry type: POLYGON
#> Dimension: XY
#> Bounding box: xmin: 913618.8 ymin: 855730.2 xmax: 1221859 ymax: 1145054
#> Projected CRS: NAD83 / Alaska Albers
#> # A tibble: 149 × 3
#> log_trend stock geometry
#> * <dbl> <fct> <POLYGON [m]>
#> 1 NA 8 ((1035002 1054710, 1035002 1054542, 1035002 1053542, 1035002…
#> 2 -0.282 8 ((1037002 1039492, 1037006 1039490, 1037017 1039492, 1037035…
#> 3 -0.00121 8 ((1070158 1030216, 1070185 1030207, 1070187 1030207, 1070211…
#> 4 0.0354 8 ((1054906 1034826, 1054931 1034821, 1054936 1034822, 1055001…
#> 5 -0.0160 8 ((1025142 1056940, 1025184 1056889, 1025222 1056836, 1025256…
#> 6 0.0872 8 ((1026035 1044623, 1026037 1044605, 1026072 1044610, 1026083…
#> 7 -0.266 8 ((1100345 1060709, 1100287 1060706, 1100228 1060706, 1100170…
#> 8 0.0743 8 ((1030247 1029637, 1030248 1029637, 1030265 1029642, 1030328…
#> 9 NA 8 ((1043093 1020553, 1043097 1020550, 1043101 1020550, 1043166…
#> 10 -0.00961 8 ((1116002 1024542, 1116002 1023542, 1116002 1022542, 1116002…
#> # ℹ 139 more rowsWe can learn more about the data by running
help("seal", "spmodel").
We can visualize the distribution of log seal trends in the
seal data by running
ggplot(seal, aes(fill = log_trend)) +
geom_sf(size = 0.75) +
scale_fill_viridis_c() +
theme_bw(base_size = 14) 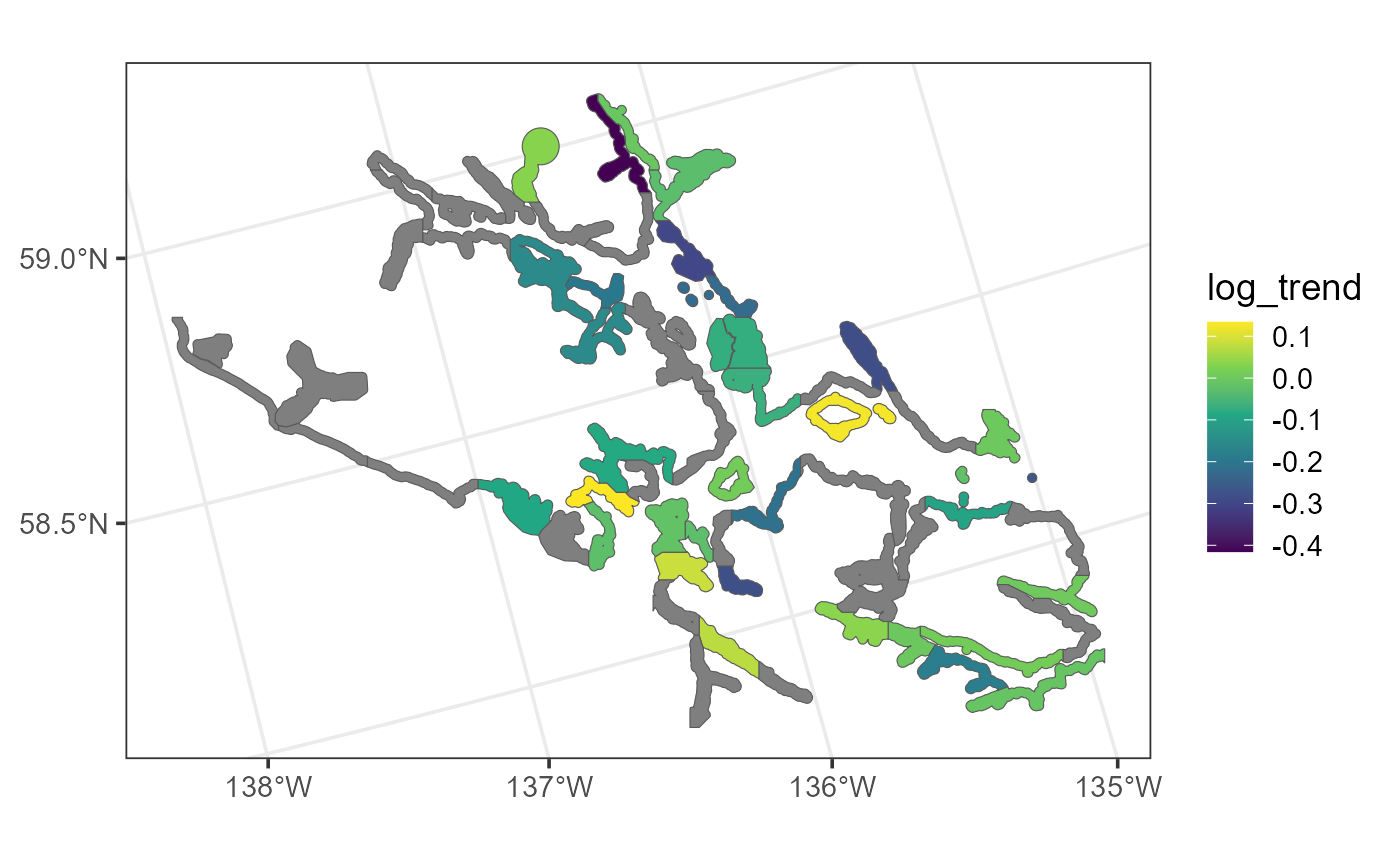
Distribution of log seal trends in the seal data. Polygons are gray if seal trends are missing.
Log trends can be viewed interactively in mapview by
running
mapview(seal, zcol = "log_trend")The gray polygons denote areas where the log trend is missing. These missing areas need to be kept in the data while fitting the model to preserve the overall neighborhood structure.
We estimate parameters of a spatial autoregressive model for log seal
trends (log_trend) using an intercept-only model with a
conditional autoregressive (CAR) spatial covariance by running
sealmod <- spautor(log_trend ~ 1, seal, spcov_type = "car")If a weight matrix is not provided to spautor(), it is
calculated internally using queen contiguity. Recall queen contiguity
defines two observations as neighbors if they share at least one common
boundary. If at least one observation has no neighbors, the
extra parameter is estimated, which quantifies variability
among observations without neighbors. By default, spautor()
uses row standardization (Ver Hoef et al.
2018) and assumes an independent error variance (ie)
of zero.
We summarize, tidy, glance at, and augment the fitted model by running
summary(sealmod)#>
#> Call:
#> spautor(formula = log_trend ~ 1, data = seal, spcov_type = "car")
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -0.402385 -0.073439 0.004368 0.073465 0.848290
#>
#> Coefficients (fixed):
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -0.01306 0.02004 -0.652 0.514
#>
#> Coefficients (car spatial covariance):
#> de range extra
#> 0.04936 0.56167 0.01750
tidy(sealmod)#> # A tibble: 1 × 5
#> term estimate std.error statistic p.value
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 (Intercept) -0.0131 0.0200 -0.652 0.514
glance(sealmod)#> # A tibble: 1 × 10
#> n p npar value AIC AICc BIC logLik deviance pseudo.r.squared
#> <int> <dbl> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 94 1 3 -74.5 -68.5 -68.3 -60.9 37.3 92.9 0
augment(sealmod)#> Simple feature collection with 94 features and 6 fields
#> Geometry type: POLYGON
#> Dimension: XY
#> Bounding box: xmin: 980001.5 ymin: 855730.2 xmax: 1221859 ymax: 1145054
#> Projected CRS: NAD83 / Alaska Albers
#> # A tibble: 94 × 7
#> log_trend .fitted .resid .hat .cooksd .std.resid
#> * <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 -0.282 -0.0131 -0.269 0.00579 0.00705 -1.10
#> 2 -0.00121 -0.0131 0.0118 0.0156 0.000527 0.182
#> 3 0.0354 -0.0131 0.0485 0.0103 0.00100 0.310
#> 4 -0.0160 -0.0131 -0.00295 0.00749 0.0000640 -0.0921
#> 5 0.0872 -0.0131 0.100 0.00939 0.00361 0.617
#> 6 -0.266 -0.0131 -0.253 0.0229 0.0877 -1.93
#> 7 0.0743 -0.0131 0.0873 0.0171 0.00620 0.597
#> 8 -0.00961 -0.0131 0.00345 0.00909 0.000254 0.166
#> 9 -0.182 -0.0131 -0.169 0.00785 0.00916 -1.08
#> 10 0.00351 -0.0131 0.0166 0.0111 0.000144 0.113
#> # ℹ 84 more rows
#> # ℹ 1 more variable: geometry <POLYGON [m]>Note that for spautor() models, the ie
spatial covariance parameter is assumed zero by default (and omitted
from the summary() output). This default behavior can be
overridden by specifying ie in the
spcov_initial argument to spautor(). Also note
that the pseudo R-squared is zero because there are no explanatory
variables in the model (i.e., it is an intercept-only model).
Prediction
In this section, we show how to use predict() to perform
spatial prediction (also called Kriging) in spmodel. We
will fit a model using the point-referenced sulfate data,
an sf object that contains sulfate measurements in the
conterminous United States, and make predictions for each location in
the point-referenced sulfate_preds data, an sf
object that contains locations in the conterminous United States at
which to predict sulfate.
We first visualize the distribution of the sulfate data by running
ggplot(sulfate, aes(color = sulfate)) +
geom_sf(size = 2.5) +
scale_color_viridis_c(limits = c(0, 45)) +
theme_gray(base_size = 18)We then fit a spatial linear model for sulfate using an intercept-only model with a spherical spatial covariance function by running
sulfmod <- splm(sulfate ~ 1, sulfate, spcov_type = "spherical")Then we obtain best linear unbiased predictions (Kriging predictions)
using predict(), where the newdata argument
contains the locations at which to predict, storing them as a new
variable in sulfate_preds called preds:
sulfate_preds$preds <- predict(sulfmod, newdata = sulfate_preds)We can visualize the model predictions by running
ggplot(sulfate_preds, aes(color = preds)) +
geom_sf(size = 2.5) +
scale_color_viridis_c(limits = c(0, 45)) +
theme_gray(base_size = 18)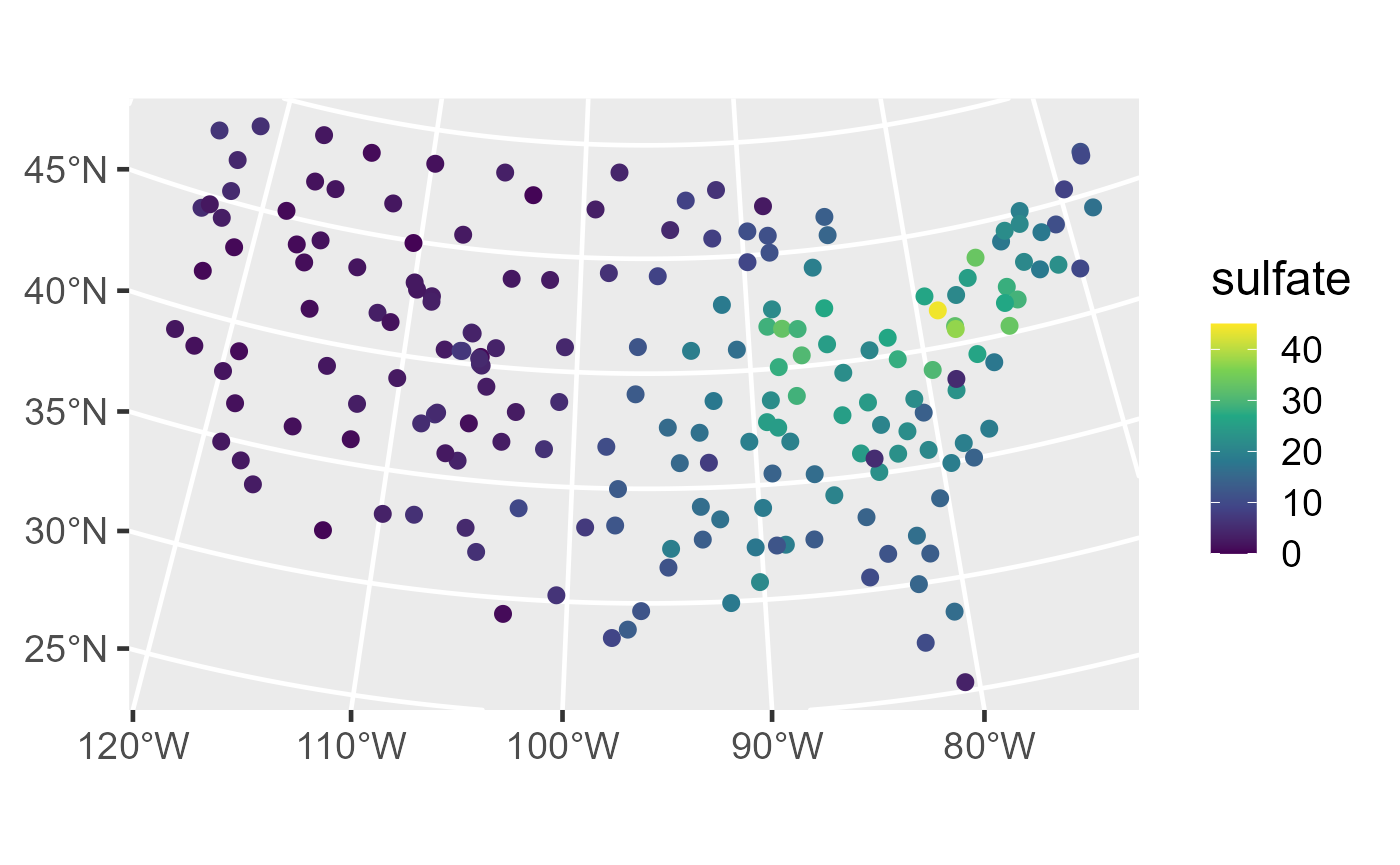
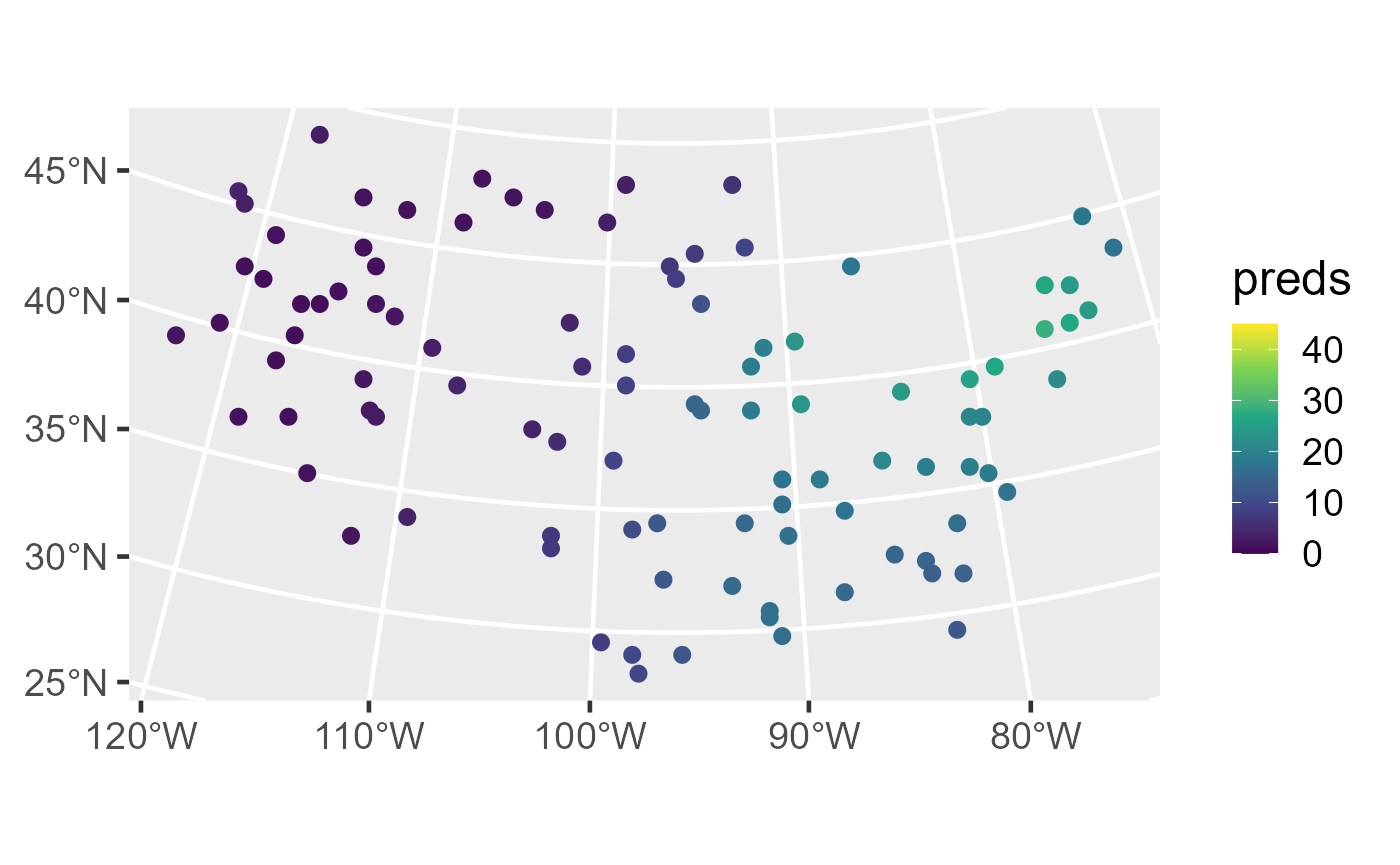
Distribution of observed sulfate (left) and sulfate predictions (right) in the conterminous United States.
Before making predictions, it is important to properly specify the
newdata object. If explanatory variables were used to fit
the model, the same explanatory variables must be included in
newdata with the same names they have in data.
Additionally, if an explanatory variable is categorical or a factor, the
values of this variable in newdata must also be values in
data (e.g., if a categorical variable with values
"A", and "B" was used to fit the model, the
corresponding variable in newdata cannot have a value
"C"). If data is a data.frame,
coordinates must be included in newdata with the same names
as they have in data. If data is an
sf object, coordinates must be included in
newdata with the same geometry name as they have in
data. When using projected coordinates, the projection for
newdata should be the same as the projection for
data.
Prediction standard errors are returned by setting the
se.fit argument to TRUE:
predict(sulfmod, newdata = sulfate_preds, se.fit = TRUE)The interval argument determines the type of interval
returned. If interval is "none" (the default),
no intervals are returned. If interval is
"prediction", 100 * level% prediction
intervals are returned (the default is 95% prediction intervals):
predict(sulfmod, newdata = sulfate_preds, interval = "prediction")If interval is "confidence", the
predictions are instead the estimated mean given each observation’s
explanatory variable values. The corresponding 100 * level%
confidence intervals are returned:
predict(sulfmod, newdata = sulfate_preds, interval = "confidence")Previously we used the augment() function to augment
data with model diagnostics. We can also use
augment() as an alternative to predict() to
augment newdata with predictions, standard errors, and
intervals. We remove the model predictions from
sulfate_preds before showing how augment() is
used to obtain the same predictions by running
sulfate_preds$preds <- NULLWe then view the first few rows of sulfate_preds
augmented with 90% prediction intervals by running
augment(sulfmod, newdata = sulfate_preds, interval = "prediction", level = 0.90)#> Simple feature collection with 100 features and 3 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: -2283774 ymin: 582930.5 xmax: 1985906 ymax: 3037173
#> Projected CRS: NAD83 / Conus Albers
#> # A tibble: 100 × 4
#> .fitted .lower .upper geometry
#> * <dbl> <dbl> <dbl> <POINT [m]>
#> 1 1.40 -5.33 8.14 (-1771413 1752976)
#> 2 24.5 18.2 30.8 (1018112 1867127)
#> 3 8.99 2.36 15.6 (-291256.8 1553212)
#> 4 16.4 9.92 23.0 (1274293 1267835)
#> 5 4.91 -1.56 11.4 (-547437.6 1638825)
#> 6 26.7 20.4 33.0 (1445080 1981278)
#> 7 3.00 -3.65 9.66 (-1629090 3037173)
#> 8 14.3 7.97 20.6 (1302757 1039534)
#> 9 1.49 -5.08 8.06 (-1429838 2523494)
#> 10 14.4 7.97 20.8 (1131970 1096609)
#> # ℹ 90 more rowsHere, .fitted represents the predictions.
An alternative (but equivalent) approach can be used for model
fitting and prediction that circumvents the need to keep
data and newdata as separate objects. Suppose
that observations requiring prediction are stored in data
as missing (NA) values. We can add a column of missing
values to sulfate_preds and then bind it together with
sulfate by running
sulfate_preds$sulfate <- NA
sulfate_with_NA <- rbind(sulfate, sulfate_preds)We can then fit a spatial linear model by running
sulfmod_with_NA <- splm(sulfate ~ 1, sulfate_with_NA, "spherical")The missing values are ignored for model-fitting but stored in
sulfmod_with_NA as newdata:
sulfmod_with_NA$newdata#> Simple feature collection with 100 features and 1 field
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: -2283774 ymin: 582930.5 xmax: 1985906 ymax: 3037173
#> Projected CRS: NAD83 / Conus Albers
#> # A tibble: 100 × 2
#> sulfate geometry
#> <dbl> <POINT [m]>
#> 1 NA (-1771413 1752976)
#> 2 NA (1018112 1867127)
#> 3 NA (-291256.8 1553212)
#> 4 NA (1274293 1267835)
#> 5 NA (-547437.6 1638825)
#> 6 NA (1445080 1981278)
#> 7 NA (-1629090 3037173)
#> 8 NA (1302757 1039534)
#> 9 NA (-1429838 2523494)
#> 10 NA (1131970 1096609)
#> # ℹ 90 more rowsWe can then predict the missing values by running
predict(sulfmod_with_NA)The call to predict() finds in
sulfmod_with_NA the newdata object and is
equivalent to
predict(sulfmod_with_NA, newdata = sulfmod_with_NA$newdata)We can also use augment() to make the predictions on the
data set with missing values by running
augment(sulfmod_with_NA, newdata = sulfmod_with_NA$newdata)#> Simple feature collection with 100 features and 2 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: -2283774 ymin: 582930.5 xmax: 1985906 ymax: 3037173
#> Projected CRS: NAD83 / Conus Albers
#> # A tibble: 100 × 3
#> sulfate .fitted geometry
#> * <dbl> <dbl> <POINT [m]>
#> 1 NA 1.40 (-1771413 1752976)
#> 2 NA 24.5 (1018112 1867127)
#> 3 NA 8.99 (-291256.8 1553212)
#> 4 NA 16.4 (1274293 1267835)
#> 5 NA 4.91 (-547437.6 1638825)
#> 6 NA 26.7 (1445080 1981278)
#> 7 NA 3.00 (-1629090 3037173)
#> 8 NA 14.3 (1302757 1039534)
#> 9 NA 1.49 (-1429838 2523494)
#> 10 NA 14.4 (1131970 1096609)
#> # ℹ 90 more rowsUnlike predict(), augment() explicitly
requires the newdata argument be specified in order to
obtain predictions. Omitting newdata (e.g., running
augment(sulfmod_with_NA)) returns model diagnostics, not
predictions.
For areal data models fit with spautor(), predictions
cannot be computed at locations that were not incorporated in the
neighborhood structure used to fit the model. Thus, predictions are only
possible for observations in data whose response values are
missing (NA), as their locations are incorporated into the
neighborhood structure. For example, we make predictions of log seal
trends at the missing polygons by running
predict(sealmod)We can also use augment() to make the predictions:
augment(sealmod, newdata = sealmod$newdata)#> Simple feature collection with 55 features and 3 fields
#> Geometry type: POLYGON
#> Dimension: XY
#> Bounding box: xmin: 913618.8 ymin: 866541.5 xmax: 1214641 ymax: 1132682
#> Projected CRS: NAD83 / Alaska Albers
#> # A tibble: 55 × 4
#> log_trend stock .fitted geometry
#> * <dbl> <fct> <dbl> <POLYGON [m]>
#> 1 NA 8 -0.106 ((1035002 1054710, 1035002 1054542, 1035002 1053542…
#> 2 NA 8 0.0359 ((1043093 1020553, 1043097 1020550, 1043101 1020550…
#> 3 NA 8 -0.0303 ((1099737 1054310, 1099752 1054262, 1099788 1054278…
#> 4 NA 8 0.00167 ((1099002 1036542, 1099134 1036462, 1099139 1036431…
#> 5 NA 8 -0.0360 ((1076902 1053189, 1076912 1053179, 1076931 1053179…
#> 6 NA 8 -0.0133 ((1070501 1046969, 1070317 1046598, 1070308 1046542…
#> 7 NA 8 -0.0704 ((1072995 1054942, 1072996 1054910, 1072997 1054878…
#> 8 NA 8 -0.0159 ((960001.5 1127667, 960110.8 1127542, 960144.1 1127…
#> 9 NA 8 -0.0558 ((1031308 1079817, 1031293 1079754, 1031289 1079741…
#> 10 NA 8 -0.0296 ((998923.7 1053647, 998922.5 1053609, 998950 105363…
#> # ℹ 45 more rowsAdvanced Features
spmodel offers several advanced features for fitting
spatial linear models. We briefly discuss some of these features next
using the moss data, the moose data, and some
simulated data. Technical details for each advanced feature can be seen
by running vignette("technical", "spmodel").
Fixing Spatial Covariance Parameters
We may desire to fix specific spatial covariance parameters at a
particular value. Perhaps some parameter value is known, for example. Or
perhaps we want to compare nested models where a reduced model uses a
fixed parameter value while the full model estimates the parameter.
Fixing spatial covariance parameters while fitting a model is possible
using the spcov_initial argument to splm() and
spautor(). The spcov_initial argument takes an
spcov_initial object (run
help("spcov_initial", "spmodel") for more).
spcov_initial objects can also be used to specify initial
values used during optimization, even if they are not assumed to be
fixed. By default, spmodel uses a grid search to find
suitable initial values to use during optimization.
As an example, suppose our goal is to compare a model with an
exponential covariance with dependent error variance, independent error
variance, and range parameters to a model that assumes the independent
random error variance parameter (nugget) is zero. First, the
spcov_initial object is specified for the latter model:
init <- spcov_initial("exponential", ie = 0, known = "ie")
print(init)#> $initial
#> ie
#> 0
#>
#> $is_known
#> ie
#> TRUE
#>
#> attr(,"class")
#> [1] "exponential"The init output shows that the ie parameter
has an initial value of zero that is assumed to be known. Next the model
is fit:
spmod_red <- splm(log_Zn ~ log_dist2road, moss, spcov_initial = init)Notice that because the spcov_initial object contains
information about the spatial covariance type, the
spcov_type argument is not required when
spcov_initial is provided. We can use
glances() to glance at both models:
glances(spmod, spmod_red)#> # A tibble: 2 × 11
#> model n p npar value AIC AICc BIC logLik deviance
#> <chr> <int> <dbl> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 spmod 365 2 3 367. 373. 373. 385. -184. 363.
#> 2 spmod_red 365 2 2 378. 382. 382. 390. -189. 374.
#> # ℹ 1 more variable: pseudo.r.squared <dbl>The lower AIC and AICc of the full model compared to the reduced
model indicates that the independent random error variance is important
to the model. A likelihood ratio test comparing the full and reduced
models is also possible using anova().
Another application of fixing spatial covariance parameters involves calculating their profile likelihood confidence intervals (Box and Cox 1964). Before calculating a profile likelihood confidence interval for \(\boldsymbol{\Theta}_i\), the \(i\)th element of a general parameter vector \(\boldsymbol{\Theta}\), it is necessary to obtain \(-2\ell(\hat{\boldsymbol{\Theta}})\), minus twice the log-likelihood evaluated at the estimated parameter vector, \(\hat{\boldsymbol{\Theta}}\). Then a \((1 - \alpha)\)% profile likelihood confidence interval is the set of values for \(\boldsymbol{\Theta}_i\) such that \(2\ell(\hat{\boldsymbol{\Theta}}) - 2\ell(\hat{\boldsymbol{\Theta}}_{-i}) \leq \chi^2_{1, 1 - \alpha}\), where \(\ell(\hat{\boldsymbol{\Theta}}_{-i})\) is the value of the log-likelihood maximized after fixing \(\boldsymbol{\Theta}_i\) and optimizing over the remaining parameters, \(\boldsymbol{\Theta}_{-i}\), and \(\chi^2_{1, 1 - \alpha}\) is the \(1 - \alpha\) quantile of a chi-squared distribution with one degree of freedom. The result follows from inverting a likelihood ratio test comparing the full model to a reduced model that fixes the value of \(\boldsymbol{\Theta}_i\). Because this approach requires refitting the model many times for different fixed values of \(\boldsymbol{\Theta}_i\), it can be computationally intensive. This approached can be generalized to yield joint profile likelihood confidence intervals cases when \(i\) has dimension greater than one.
Fitting and Predicting for Multiple Models
Fitting multiple models is possible with a single call to
splm() or spautor() when
spcov_type is a vector with length greater than one or
spcov_initial is a list (with length greater than one) of
spcov_initial objects. We fit three separate spatial linear
models using the exponential spatial covariance, spherical spatial
covariance, and no spatial covariance by running
spmods is list whose names indicate the spatial
covariance type and whose values indicate the model fit for that spatial
covariance type. Generic functions can be called individually for each
list element. For example, we can augment the the model fit using the
exponential covariance function with diagnostics by running
augment(spmods$exponential)#> Simple feature collection with 197 features and 6 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: -2292550 ymin: 386181.1 xmax: 2173345 ymax: 3090370
#> Projected CRS: NAD83 / Conus Albers
#> # A tibble: 197 × 7
#> sulfate .fitted .resid .hat .cooksd .std.resid geometry
#> * <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <POINT [m]>
#> 1 12.9 5.92 7.00 0.00334 0.00161 -0.694 (817738.8 1080571)
#> 2 20.2 5.92 14.2 0.00256 0.00192 0.865 (914593.6 1295545)
#> 3 16.8 5.92 10.9 0.00259 0.000395 0.390 (359574.1 1178228)
#> 4 16.2 5.92 10.3 0.00239 0.000363 0.390 (265331.9 1239089)
#> 5 7.86 5.92 1.93 0.00202 0.00871 -2.07 (304528.8 1453636)
#> 6 15.4 5.92 9.43 0.00201 0.000240 0.345 (162932.8 1451625)
#> 7 0.986 5.92 -4.94 0.00380 0.000966 -0.503 (-1437776 1568022)
#> 8 0.425 5.92 -5.50 0.0138 0.00584 -0.646 (-1572878 1125529)
#> 9 3.58 5.92 -2.34 0.00673 0.0000148 -0.0467 (-1282009 1204889)
#> 10 2.38 5.92 -3.54 0.0123 0.0000139 -0.0335 (-1972775 1464991)
#> # ℹ 187 more rowsOr we can find the AIC of the model fit using the spherical covariance function by running
AIC(spmods$spherical)#> [1] 1143.202The glances() and predict() functions can
work directly with spmods, calling glances()
and predict() on each list element and then organizing the
results. We glance at each fitted model object by running
glances(spmods)#> # A tibble: 3 × 11
#> model n p npar value AIC AICc BIC logLik deviance
#> <chr> <int> <dbl> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 spherical 197 1 3 1137. 1143. 1143. 1153. -569. 196.
#> 2 exponential 197 1 3 1140. 1146. 1146. 1156. -570. 196.
#> 3 none 197 1 1 1448. 1450. 1450. 1453. -724. 196
#> # ℹ 1 more variable: pseudo.r.squared <dbl>We predict newdata separately for each fitted model
object by running
predict(spmods, newdata = sulfate_preds)Random Effects
Non-spatial random effects incorporate additional sources of
variability into model fitting. They are accommodated in
spmodel using similar syntax as for random effects in the
nlme (Pinheiro and Bates
2006) and lme4 (Bates et al.
2015) R packages. Random effects are specified
via a formula passed to the random argument. Next we show
two examples that incorporate random effects into the spatial linear
model using the moss data.
The first example explores random intercepts for the
sample variable. The sample variable indexes
each unique location, which can have replicate observations due to field
duplicates (field_dup) and lab replicates
(lab_rep). There are 365 observations in moss
at 318 unique locations, which means that 47 observations in
moss are either field duplicates or lab replicates. It is
likely that the repeated observations at a location are correlated with
one another. We can incorporate this repeated-observation correlation by
creating a random intercept for each level of sample. We
model the random intercepts for sample by running
rand1 <- splm(
log_Zn ~ log_dist2road,
moss,
spcov_type = "exponential",
random = ~ sample
)Note that ~ sample is shorthand for
(1 | sample), which is more explicit notation that
indicates random intercepts for each level of sample.
The second example adds a random intercept for year,
which creates extra correlation for observations within a year. It also
adds a random slope for log_dist2road within
year, which lets the effect of log_dist2road
vary between years. We fit this model by running
rand2 <- splm(
log_Zn ~ log_dist2road,
moss,
spcov_type = "exponential",
random = ~ sample + (log_dist2road | year)
)Note that sample + (log_dist2road | year) is shorthand
for (1 | sample) + (log_dist2road | year). If only random
slopes within a year are desired (and no random intercepts), a
- 1 is given to the relevant portion of the formula:
(log_dist2road - 1 | year). When there is more than one
term in random, each term must be surrounded by parentheses
(recall that the random intercept shorthand automatically includes
relevant parentheses). More examples of random effect syntax are
provided in the appendix.
We compare the AIC of the models by running
glances(rand1, rand2)#> # A tibble: 2 × 11
#> model n p npar value AIC AICc BIC logLik deviance
#> <chr> <int> <dbl> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 rand2 365 2 6 190. 202. 202. 225. -94.9 363.
#> 2 rand1 365 2 4 335. 343. 343. 359. -168. 363.
#> # ℹ 1 more variable: pseudo.r.squared <dbl>The rand2 model has the lowest AIC.
It is possible to fix random effect variances using the
randcov_initial argument, and randcov_initial
can also be used to set initial values for optimization.
Partition Factors
A partition factor is a variable that allows observations to be
uncorrelated when they are from different levels of the partition
factor. Partition factors are specified in spmodel by
providing a formula with a single variable to the
partition_factor argument. Suppose that for the
moss data, we want observations in different years
(year) to be uncorrelated. We fit a model that treats year
as a partition factor by running
part <- splm(
log_Zn ~ log_dist2road,
moss,
spcov_type = "exponential",
partition_factor = ~ year
)Anisotropy
An isotroptic spatial covariance function (for point-referenced data) behaves similarly in all directions (i.e., is independent of direction) as a function of distance. An anisotropic covariance function does not behave similarly in all directions as a function of distance. Consider the spatial covariance imposed by an eastward-moving wind pattern. A one-unit distance in the x-direction likely means something different than a one-unit distance in the y-direction. Below are ellipses for an isotropic and anisotropic covariance function centered at the origin (a distance of zero).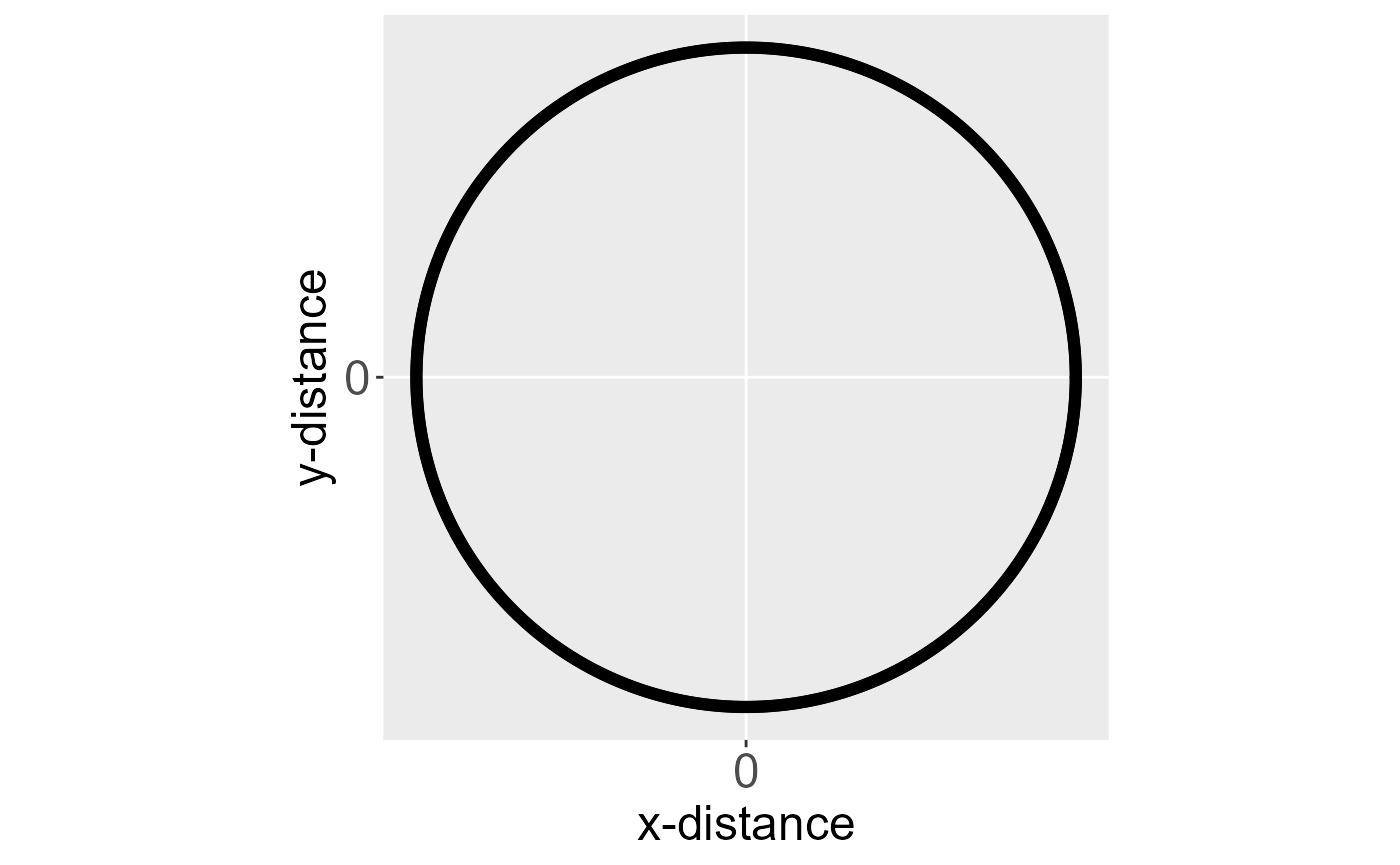
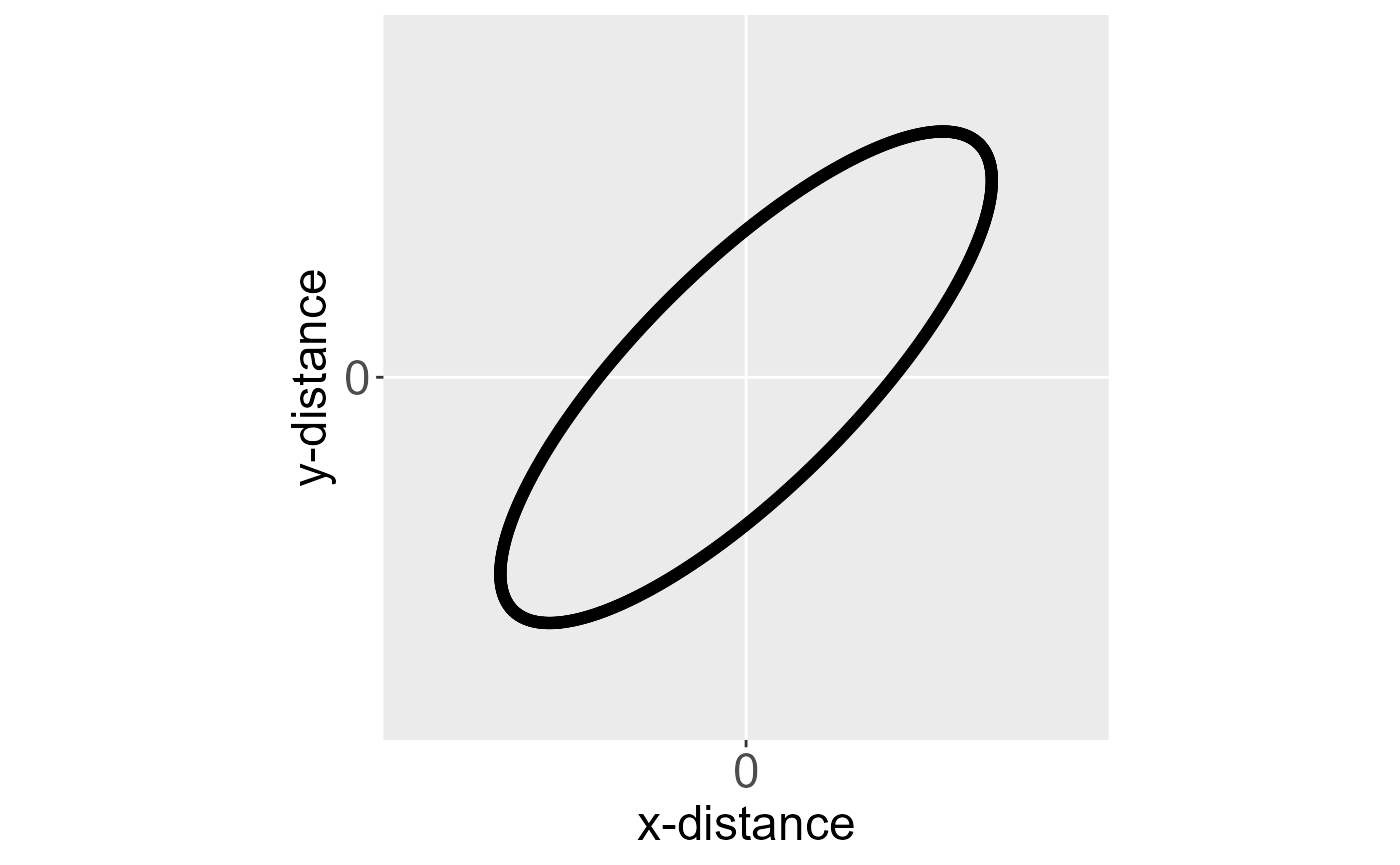
Ellipses for an isotropic (left) and anisotropic (right) covariance function centered at the origin. The black outline of each ellipse is a level curve of equal correlation.
The black outline of each ellipse is a level curve of equal correlation. The left ellipse (a circle) represents an isotropic covariance function. The distance at which the correlation between two observations lays on the level curve is the same in all directions. The right ellipse represents an anisotropic covariance function. The distance at which the correlation between two observations lays on the level curve is different in different directions.
Accounting for anisotropy involves a rotation and scaling of the
x-coordinates and y-coordinates such that the spatial covariance
function that uses these transformed distances is isotropic. We use the
anisotropy argument to splm() to fit a model
with anisotropy by running
spmod_anis <- splm(
log_Zn ~ log_dist2road,
moss,
spcov_type = "exponential",
anisotropy = TRUE
)
summary(spmod_anis)#>
#> Call:
#> splm(formula = log_Zn ~ log_dist2road, data = moss, spcov_type = "exponential",
#> anisotropy = TRUE)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -2.5279 -1.2239 -0.7202 -0.1921 1.1659
#>
#> Coefficients (fixed):
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 9.54798 0.22291 42.83 <2e-16 ***
#> log_dist2road -0.54601 0.01855 -29.44 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Pseudo R-squared: 0.7048
#>
#> Coefficients (exponential spatial covariance):
#> de ie range rotate scale
#> 3.561e-01 6.812e-02 8.732e+03 2.435e+00 4.753e-01
#> attr(,"class")
#> [1] "exponential"The rotate parameter is between zero and \(\pi\) radians and represents the angle of a
clockwise rotation of the ellipse such that the major axis of the
ellipse is the new x-axis and the minor axis of the ellipse is the new
y-axis. The scale parameter is between zero and one and
represents the ratio of the distance between the origin and the edge of
the ellipse along the minor axis to the distance between the origin and
the edge of the ellipse along the major axis. Below shows a
transformation that turns an anisotropic ellipse into an isotropic one
(i.e., a circle). This transformation requires rotating the coordinates
clockwise by rotate and then scaling them the reciprocal of
scale. The transformed coordinates are then used instead of
the original coordinates to compute distances and spatial
covariances.
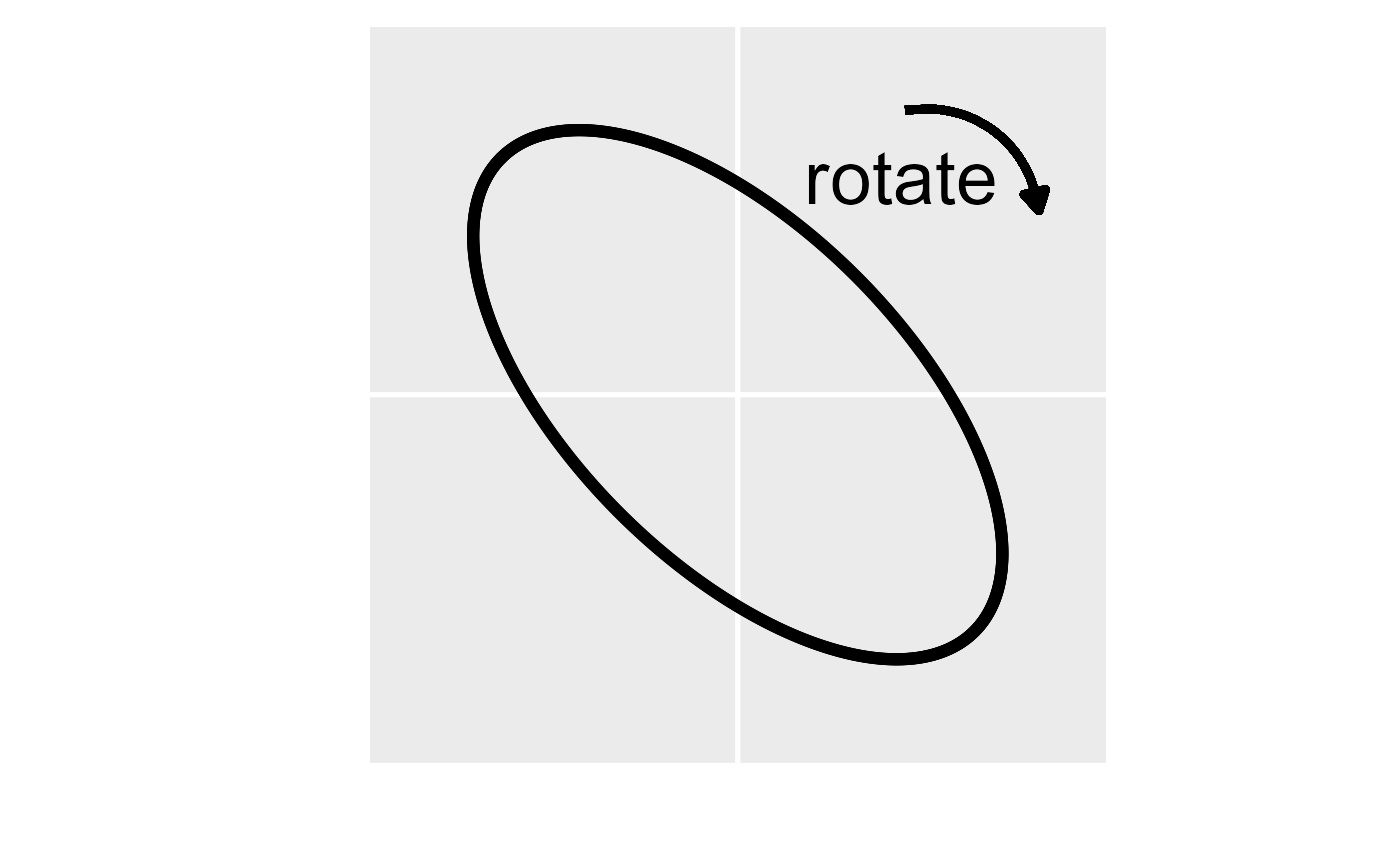
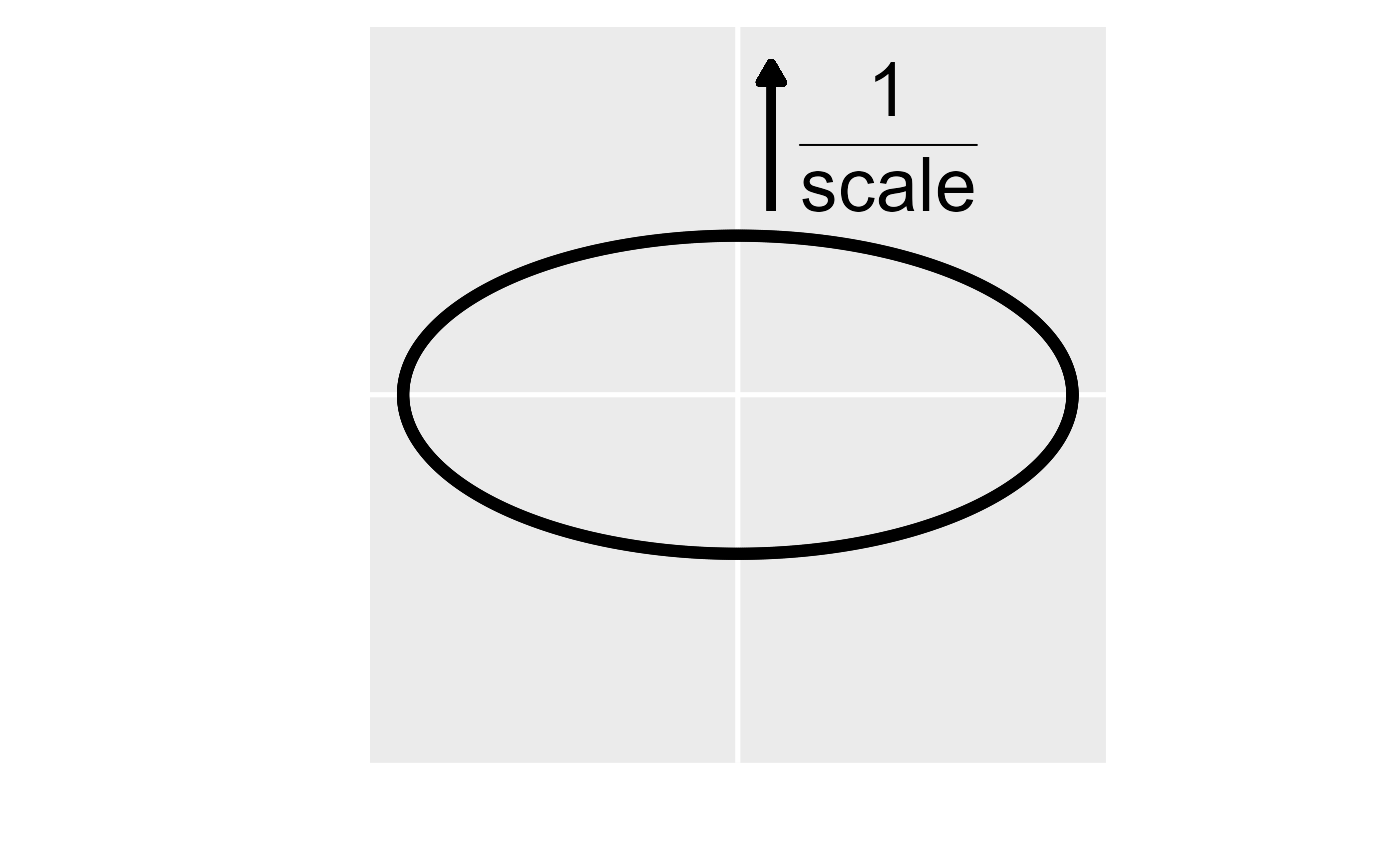
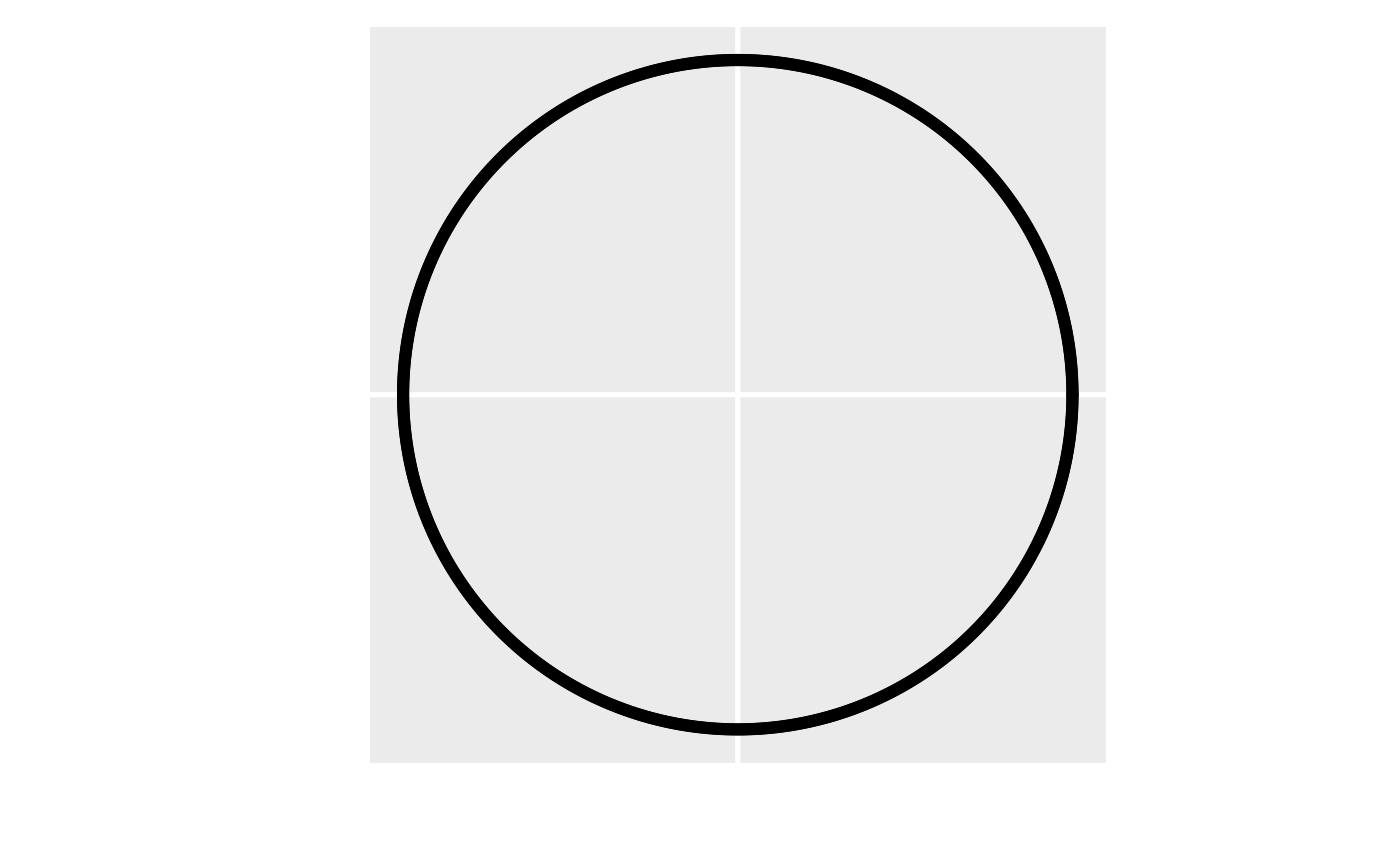
A visual representation of the anisotropy transformation. In the left
figure, the first step is to rotate the anisotropic ellipse clockwise by
the rotate parameter (here rotate is 0.75\(\pi\) radians or 135 degrees). In the
middle figure, the second step is to scale the y axis by the reciprocal
of the scale parameter (here scale is 0.5). In
the right figure, the anisotropic ellipse has been transformed into an
isotropic one (i.e., a circle). The transformed coordinates are then
used instead of the original coordinates to compute distances and
spatial covariances.
Note that specifying an initial value for rotate that is
different from zero, specifying an initial value for scale
that is different from one, or assuming either rotate or
scale are unknown in spcov_initial will cause
splm() to fit a model with anisotropy (and will override
anisotropy = FALSE). Estimating anisotropy parameters is
only possible for maximum likelihood and restricted maximum likelihood
estimation, but fixed anisotropy parameters can be accommodated for
semivariogram weighted least squares or semivariogram composite
likelihood estimation. Also note that anisotropy is not relevant for
areal data because the spatial covariance function depends on a
neighborhood structure instead of distances between points.
Simulating Spatial Data
The sprnorm() function is used to simulate normal
(Gaussian) spatial data. To use sprnorm(), the
spcov_params() function is used to create an
spcov_params object. The spcov_params()
function requires the spatial covariance type and parameter values. We
create an spcov_params object by running
sim_params <- spcov_params("exponential", de = 5, ie = 1, range = 0.5)We set a reproducible seed and then simulate data at 3000 random
locations in the unit square using the spatial covariance parameters in
sim_params by running
set.seed(0)
n <- 3000
x <- runif(n)
y <- runif(n)
sim_coords <- tibble::tibble(x, y)
sim_response <- sprnorm(sim_params, data = sim_coords, xcoord = x, ycoord = y)
sim_data <- tibble::tibble(sim_coords, sim_response)We can visualize the simulated data by running
ggplot(sim_data, aes(x = x, y = y, color = sim_response)) +
geom_point(size = 1.5) +
scale_color_viridis_c(limits = c(-7, 7)) +
theme_gray(base_size = 18)There is noticeable spatial patterning in the response variable
(sim_response). The default mean in sprnorm()
is zero for all observations, though a mean vector can be provided using
the mean argument. The default number of samples generated
in sprnorm() is one, though this can be changed using the
samples argument. Because sim_data is a
tibble (data.frame) and not an sf
object, the columns in sim_data representing the
x-coordinates and y-coordinates must be provided to
sprnorm().
Note that the output from coef(object, type = "spcov")
is a spcov_params object. This is useful if we want to
simulate data given the estimated spatial covariance parameters from a
fitted model. Random effects are incorporated into simulation via the
randcov_params argument.
Big Data
The computational cost associated with model fitting is exponential
in the sample size for all estimation methods. For maximum likelihood
and restricted maximum likelihood, the computational cost of estimating
\(\boldsymbol{\theta}\) is cubic. For
semivariogram weighted least squares and semivariogram composite
likelihood, the computational cost of estimating \(\boldsymbol{\theta}\) is quadratic. The
computational cost associated with estimating \(\boldsymbol{\beta}\) and prediction is
cubic, regardless of estimation method. Typically, samples sizes
approaching 10,000 make the computational cost of model fitting and
prediction infeasible, which necessitates the use of big data methods.
spmodel offers big data methods for model fitting of
point-referenced data via the local argument to
splm(). The method is capable of quickly fitting models
with hundreds of thousands of observations. Because of the neighborhood
structure of areal data, the big data methods used for point-referenced
data do not apply to areal data. Thus, there is no big data method for
areal data or local argument to spautor(), so
model fitting sample sizes cannot be too large
spmodel offers big data methods for prediction of
point-referenced data or areal data via the local argument
to predict(). This method is capable of quickly predicting
hundreds of thousands of observations. To show how to use
spmodel for big data estimation and prediction, we use the
sim_data data from the previous section. Because
sim_data is a tibble (data.frame)
and not an sf object, the columns in data
representing the x-coordinates and y-coordinates must be explicitly
provided to splm().
Next we briefly discuss model fitting and prediction for big data in
spmodel, but further details are provided by Ver Hoef et al. (2023).
Model Fitting
spmodel uses a “local spatial indexing” (SPIN) approach
for big data model fitting of point-referenced data. Observations are
first assigned an index. Then for the purposes of model fitting,
observations with different indexes are assumed uncorrelated. Assuming
observations with different indexes are uncorrelated induces sparsity in
the covariance matrix, which greatly reduces the computational time of
operations that involve the covariance matrix.
The local argument to splm() controls the
big data options. local is a list with several arguments.
The arguments to the local list control the method used to
assign the indexes, the number of observations with the same index, the
number of unique indexes, variance adjustments to the covariance matrix
of \(\hat{\boldsymbol{\beta}}\),
whether or not to use parallel processing, and if parallel processing is
used, the number of cores.
Big data are most simply accommodated by setting local
to TRUE. This is shorthand for
local = list(method = "kmeans", size = 100, var_adjust = "theoretical", parallel = FALSE),
which assigns observations to index groups based on k-means clustering (MacQueen 1967), ensures each index group has approximately 100 observations, uses the theoretically-correct variance adjustment, and does not use parallel processing.
local1 <- splm(sim_response ~ 1, sim_data, spcov_type = "exponential",
xcoord = x, ycoord = y, local = TRUE)
summary(local1)#>
#> Call:
#> splm(formula = sim_response ~ 1, data = sim_data, spcov_type = "exponential",
#> xcoord = x, ycoord = y, local = TRUE)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -4.9414 -1.2573 -0.0527 1.3783 6.6322
#>
#> Coefficients (fixed):
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -1.1149 0.6416 -1.738 0.0823 .
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Coefficients (exponential spatial covariance):
#> de ie range
#> 2.3112 1.0260 0.2395Instead of using local = TRUE, we can explicitly set
local. For example, we can fit a model using random
assignment with 60 groups, use the pooled variance adjustment, and use
parallel processing with two cores by running
local2_list <- list(method = "random", groups = 60, var_adjust = "pooled",
parallel = TRUE, ncores = 2)
local2 <- splm(sim_response ~ 1, sim_data, spcov_type = "exponential",
xcoord = x, ycoord = y, local = local2_list)Likelihood-based statistics like AIC(),
AICc(), BIC(), logLik(), and
deviance() should not be used to compare a model fit with a
big data approximation to a model fit without a big data
approximation.
Prediction
For point-referenced data, spmodel uses a “local
neighborhood” approach for big data prediction. Each prediction is
computed using a subset of the observed data instead of all of the
observed data. Before further discussing big data prediction, we
simulate 1000 locations in the unit square requiring prediction:
The local argument to predict() controls
the big data options. local is a list with several
arguments. The arguments to the local list control the
method used to subset the observed data, the number of observations in
each subset, whether or not to use parallel processing, and if parallel
processing is used, the number of cores.
The simplest way to accommodate big data prediction is to set
local to TRUE. This is shorthand for
local = list(method = "covariance", size = 100, parallel = FALSE),
which implies that, for each location requiring prediction, only the 100
observations in the data most correlated with it are used in the
computation and parallel processing is not used. Using the
local1 fitted model, we store these predictions as a
variable called preds in the sim_preds data by
running
sim_preds$preds <- predict(local1, newdata = sim_preds, local = TRUE)The predictions are visualized by running
ggplot(sim_preds, aes(x = x, y = y, color = preds)) +
geom_point(size = 1.5) +
scale_color_viridis_c(limits = c(-7, 7)) +
theme_gray(base_size = 18)They display a similar pattern as the observed data.
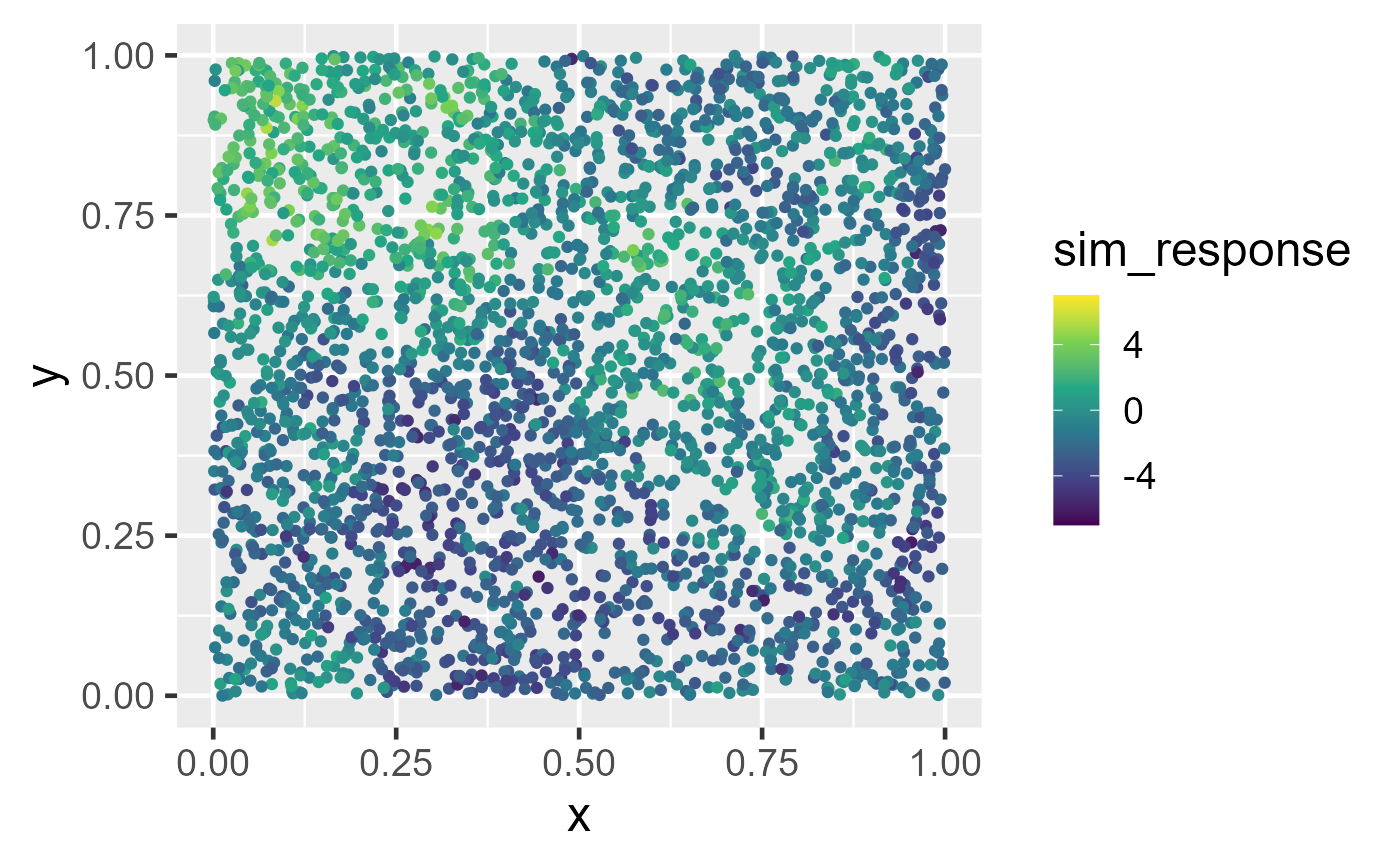
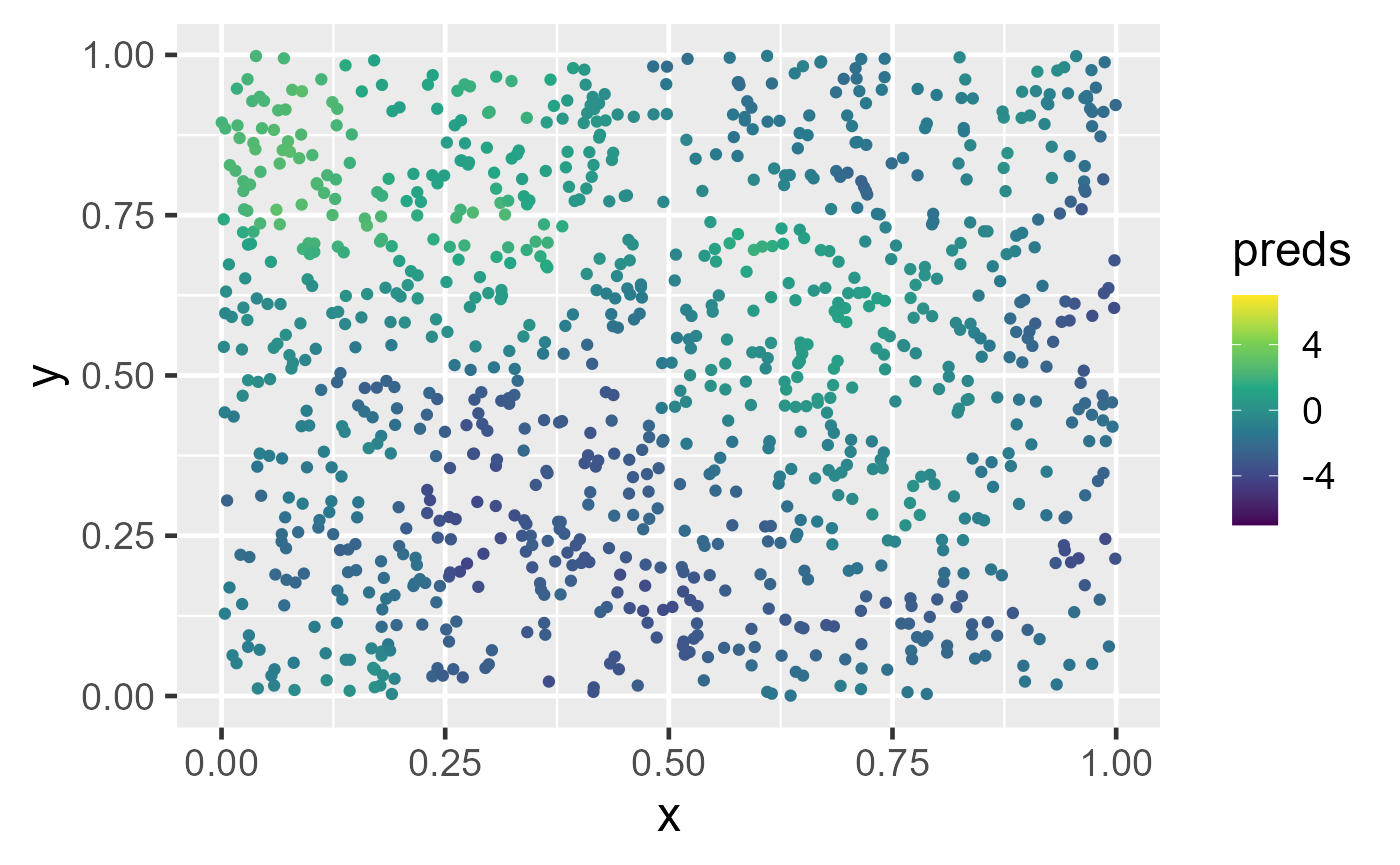
Observed data and big data predictions at unobserved locations. In the left figure, spatial data are simulated in the unit square. A spatial linear model is fit using the default big data approximation for model-fitting. In the right figure, predictions are made using the fitted model and the default big data approximation for prediction.
Instead of using local = TRUE, we can explicitly set
local:
pred_list <- list(method = "distance", size = 30, parallel = TRUE, ncores = 2)
predict(local1, newdata = sim_preds, local = pred_list)This code implies that uniquely for each location requiring prediction, only the 30 observations in the data closest to it (in terms of Euclidean distance) are used in the computation and parallel processing is used with two cores.
For areal data, no local neighborhood approximation exists because of
the data’s underlying neighborhood structure. Thus, all of the data must
be used to compute predictions, and by consequence, method
and size are not components of the local list.
The only components of the local list for areal data are
parallel and ncores.
Random Forest Spatial Residual Models
Random forest spatial residual models are used for prediction. They
combine aspects of random forest prediction and spatial linear model
prediction, which can lead to significant improvements in predictive
accuracy compared to standard random forest prediction (Fox, Ver Hoef, and Olsen 2020). To fit a random
forest spatial residual model, use splmRF() (for
point-referenced data) or spautorRF() (for areal data).
These functions require at least one explanatory variable be specified,
so we add an explanatory variable called var to
sulfate and sulfate_preds for illustrative
purposes.
Then we fit a random forest spatial residual model by running
sprfmod <- splmRF(sulfate ~ var, sulfate, spcov_type = "exponential")And we make predictions by running
predict(sprfmod, newdata = sulfate_preds)Spatial Generalized Linear Models
When building spatial linear models, the response vector \(\mathbf{y}\) is typically assumed Gaussian (given \(\mathbf{X}\)). Relaxing this assumption on the distribution of \(\mathbf{y}\) yields a rich class of spatial generalized linear models that can describe binary data, proportion data, count data, and skewed data that is parameterized as \[ g(\boldsymbol{\mu}) = \boldsymbol{\eta} = \mathbf{X} \boldsymbol{\beta} + \boldsymbol{\tau} + \boldsymbol{\epsilon}, \] where \(g(\cdot)\) is called a link function, \(\boldsymbol{\mu}\) is the mean of \(\mathbf{y}\), and the remaining terms \(\mathbf{X}\), \(\boldsymbol{\beta}\), \(\boldsymbol{\tau}\), \(\boldsymbol{\epsilon}\) represent the same quantities as for the spatial linear models. The link function, \(g(\cdot)\), “links” a function of \(\boldsymbol{\mu}\) to the linear term \(\boldsymbol{\eta}\) , denoted here as \(\mathbf{X} \boldsymbol{\beta} + \boldsymbol{\tau} + \boldsymbol{\epsilon}\), which is familiar from spatial linear models. Note that the linking of \(\boldsymbol{\mu}\) to \(\boldsymbol{\eta}\) applies element-wise to each vector. Each link function \(g(\cdot)\) has a corresponding inverse link function, \(g^{-1}(\cdot)\). The inverse link function “links” a function of \(\boldsymbol{\eta}\) to \(\boldsymbol{\mu}\). Notice that for spatial generalized linear models, we are not modeling \(\mathbf{y}\) directly as we do for spatial linear models, but rather we are modeling a function of the mean of \(\mathbf{y}\). Also notice that \(\boldsymbol{\eta}\) is unconstrained but \(\boldsymbol{\mu}\) is usually constrained in some way (e.g., positive).
The model \(g(\boldsymbol{\mu}) =
\boldsymbol{\eta} = \mathbf{X} \boldsymbol{\beta} + \boldsymbol{\tau} +
\boldsymbol{\epsilon}\) is called the spatial generalized linear
model. spmodel allows fitting of spatial generalized linear
models when \(\mathbf{y}\) is a
binomial (or Bernoulli), beta, Poisson, negative binomial, gamma, or
inverse Gaussian random vector via the Laplace approximation and
restricted maximum likelihood estimation or maximum likelihood
estimation – Ver Hoef et al. (2024)
provide further details. For binomial and beta \(\mathbf{y}\), the logit link function is
defined as \(g(\boldsymbol{\mu}) =
\ln(\frac{\boldsymbol{\mu}}{1 - \boldsymbol{\mu}}) =
\boldsymbol{\eta}\), and the inverse logit link function is
defined as \(g^{-1}(\boldsymbol{\eta}) =
\frac{\exp(\boldsymbol{\eta})}{1 + \exp(\boldsymbol{\eta})} =
\boldsymbol{\mu}\). For Poisson, negative binomial, gamma, and
inverse Gaussian \(\mathbf{y}\), the
log link function is defined as \(g(\boldsymbol{\mu}) = \ln(\boldsymbol{\mu}) =
\boldsymbol{\eta}\), and the inverse log link function is defined
as \(g^{-1}(\boldsymbol{\eta}) =
\exp(\boldsymbol{\eta}) = \boldsymbol{\mu}\). Full
parameterizations of these distributions are given in the technical
details vignette, which can be viewed by running
vignette("technical", "spmodel").
All advanced features available in spmodel for spatial
linear models are also available for spatial generalized linear models.
This means that spatial generalized linear models in
spmodel can accommodate fixing spatial covariance
parameters, fitting and predicting for multiple models, random effects
(on the link scale), partition factors, anisotropy (on the link scale),
simulation, big data, and prediction.
Model Fitting
As with spatial linear models, spatial generalized linear models are
fit in spmodel for point-referenced and areal data. The
spglm() function is used to fit spatial generalized linear
models for point-referenced data, and the spgautor()
function is used to fit spatial generalized linear models for areal
data. spglm() and spgautor() share similar
syntax with splm() and spautor(),
respectively, though one additional argument is required:
-
family: the generalized linear model family (i.e., the distribution of \(\mathbf{y}\)).familycan bebinomial,beta,poisson,nbinomial,Gamma, orinverse.gaussian. -
familyuses similar syntax as thefamilyargument inglm(). - One difference between
familyinspglm()compared tofamilyinglm()is that the link function is fixed inspglm().
As with splm() and spautor(), the data
argument to spglm() and spgautor() can be an
sf object or data.frame (with appropriate
coordinate or weight matrix information). Additionally, all arguments to
splm() and spautor() are also available for
spglm() and spgautor().
Next we show the basic features and syntax of spglm()
using the moose data. We study the impact of elevation
(elev) on the presence of moose (presence)
observed at a site location in Alaska. presence equals one
if at least one moose was observed at the site and zero otherwise. We
view the first few rows of the moose data by running
moose#> Simple feature collection with 218 features and 4 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: 269085 ymin: 1416151 xmax: 419976.2 ymax: 1541763
#> Projected CRS: NAD83 / Alaska Albers
#> # A tibble: 218 × 5
#> elev strat count presence geometry
#> <dbl> <chr> <dbl> <fct> <POINT [m]>
#> 1 469. L 0 0 (293542.6 1541016)
#> 2 362. L 0 0 (298313.1 1533972)
#> 3 173. M 0 0 (281896.4 1532516)
#> 4 280. L 0 0 (298651.3 1530264)
#> 5 620. L 0 0 (311325.3 1527705)
#> 6 164. M 0 0 (291421.5 1518398)
#> 7 164. M 0 0 (287298.3 1518035)
#> 8 186. L 0 0 (279050.9 1517324)
#> 9 362. L 0 0 (346145.9 1512479)
#> 10 430. L 0 0 (321354.6 1509966)
#> # ℹ 208 more rowsWe can visualize the distribution of moose presence
(presence) by running
ggplot(moose, aes(color = presence)) +
geom_sf(size = 2) +
scale_color_viridis_d(option = "H") +
theme_gray(base_size = 14)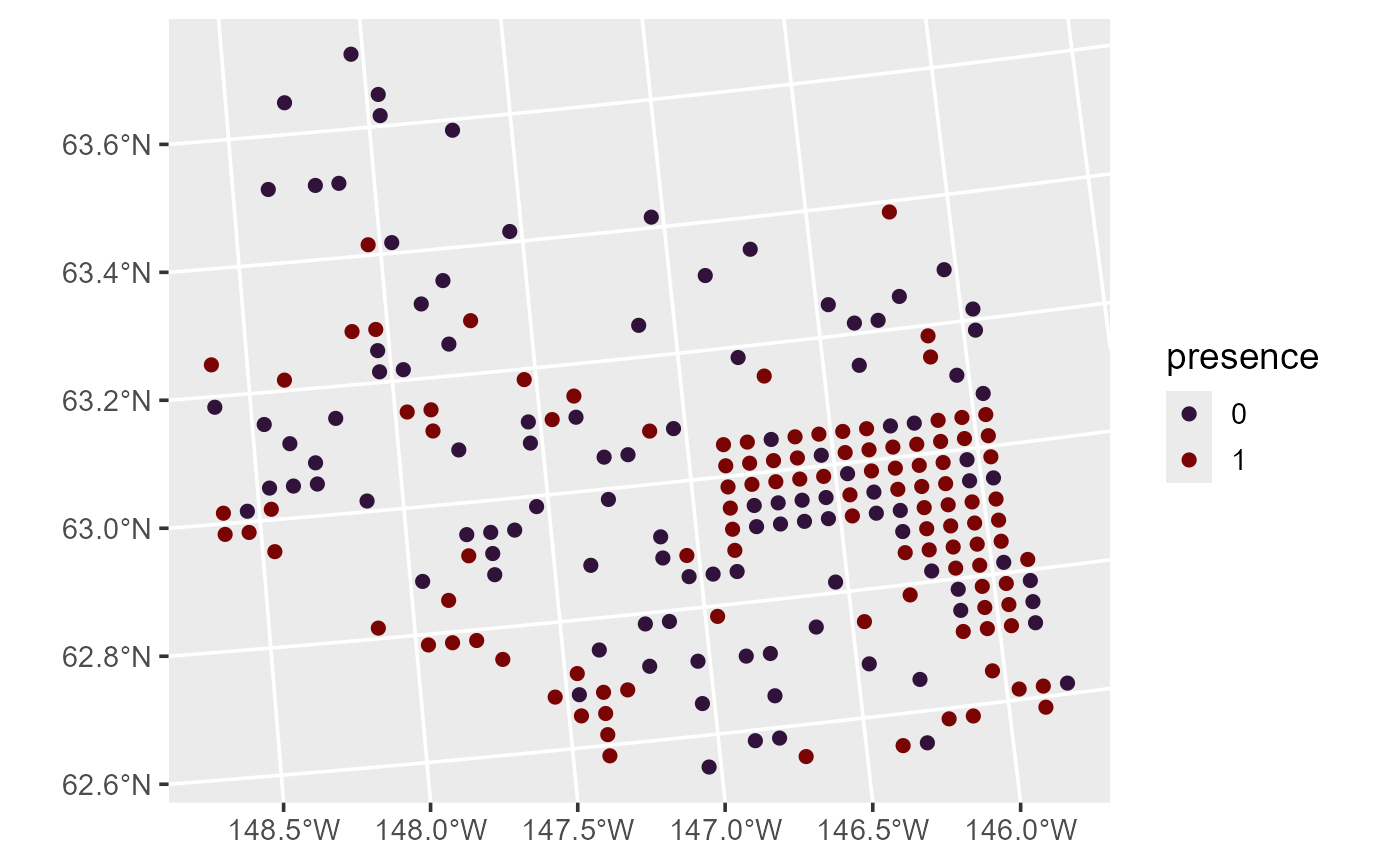
Distribution of moose presence in Alaska. presence equals one if at least one moose was observed at the site and zero otherwise.
One example of a generalized linear model is a binomial (e.g., logistic) regression model. Binomial regression models are often used to model presence data such as these. To quantify the relationship between moose presence and elevation, we fit a spatial binomial regression model (a specific spatial generalized linear model) by running
binmod <- spglm(presence ~ elev, family = "binomial",
data = moose, spcov_type = "exponential")The estimation method is specified via the estmethod
argument, which has a default value of "reml" for
restricted maximum likelihood. The other estimation method is
"ml" for maximum likelihood.
We summarize the fitted model by running
summary(binmod)#>
#> Call:
#> spglm(formula = presence ~ elev, family = "binomial", data = moose,
#> spcov_type = "exponential")
#>
#> Deviance Residuals:
#> Min 1Q Median 3Q Max
#> -1.5249 -0.8114 0.5600 0.8306 1.5757
#>
#> Coefficients (fixed):
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -0.874038 1.140953 -0.766 0.444
#> elev 0.002365 0.003184 0.743 0.458
#>
#> Pseudo R-squared: 0.00311
#>
#> Coefficients (exponential spatial covariance):
#> de ie range
#> 3.746e+00 4.392e-03 3.203e+04
#>
#> Coefficients (Dispersion for binomial family):
#> dispersion
#> 1Similar to summaries of glm() objects, summaries of
spglm() objects include the original function call, summary
statistics of the deviance residuals, and a coefficients table of fixed
effects. The logit of moose presence probability does not appear to be
related to elevation, as evidenced by the large p-value associated with
the asymptotic z-test. A pseudo r-squared is also returned, which
quantifies the proportion of variability explained by the fixed effects.
The spatial covariance parameters and dispersion parameter are also
returned. The dispersion parameter is estimated in some spatial
generalized linear models and changes the mean-variance relationship of
\(\mathbf{y}\). For binomial regression
models, the dispersion parameter is not estimated and is always fixed at
one.
We tidy, glance, and augment the fitted model by running
tidy(binmod)#> # A tibble: 2 × 5
#> term estimate std.error statistic p.value
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 (Intercept) -0.874 1.14 -0.766 0.444
#> 2 elev 0.00237 0.00318 0.743 0.458
glance(binmod)#> # A tibble: 1 × 10
#> n p npar value AIC AICc BIC logLik deviance pseudo.r.squared
#> <int> <dbl> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 218 2 3 692. 698. 698. 708. -346. 190. 0.00311
augment(binmod)#> Simple feature collection with 218 features and 7 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: 269085 ymin: 1416151 xmax: 419057.4 ymax: 1541016
#> Projected CRS: NAD83 / Alaska Albers
#> # A tibble: 218 × 8
#> presence elev .fitted .resid .hat .cooksd .std.resid
#> * <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 0 469. -1.73 -0.571 0.0500 0.00904 -0.586
#> 2 0 362. -2.12 -0.477 0.0168 0.00198 -0.481
#> 3 0 173. -2.15 -0.468 0.00213 0.000235 -0.469
#> 4 0 280. -2.27 -0.444 0.00616 0.000615 -0.445
#> 5 0 620. -1.40 -0.664 0.136 0.0402 -0.714
#> 6 0 164. -1.90 -0.528 0.00260 0.000364 -0.528
#> 7 0 164. -1.86 -0.538 0.00269 0.000392 -0.539
#> 8 0 186. -1.61 -0.603 0.00332 0.000607 -0.604
#> 9 0 362. -1.60 -0.606 0.0245 0.00474 -0.614
#> 10 0 430. -1.24 -0.714 0.0528 0.0150 -0.734
#> # ℹ 208 more rows
#> # ℹ 1 more variable: geometry <POINT [m]>Prediction
For spatial generalized linear models, we are predicting the mean of
the process generating the observation (which includes the spatial
effects) rather than the observation itself. We make predictions of
moose presence probability at the locations in moose_preds
by running
moose_preds$preds <- predict(binmod, newdata = moose_preds, type = "response")The type argument specifies whether predictions are returned on the link or response (inverse link) scale.
We visualize these predictions by running
ggplot(moose_preds, aes(color = preds)) +
geom_sf(size = 2) +
scale_color_viridis_c(limits = c(0, 1), option = "H") +
theme_gray(base_size = 14)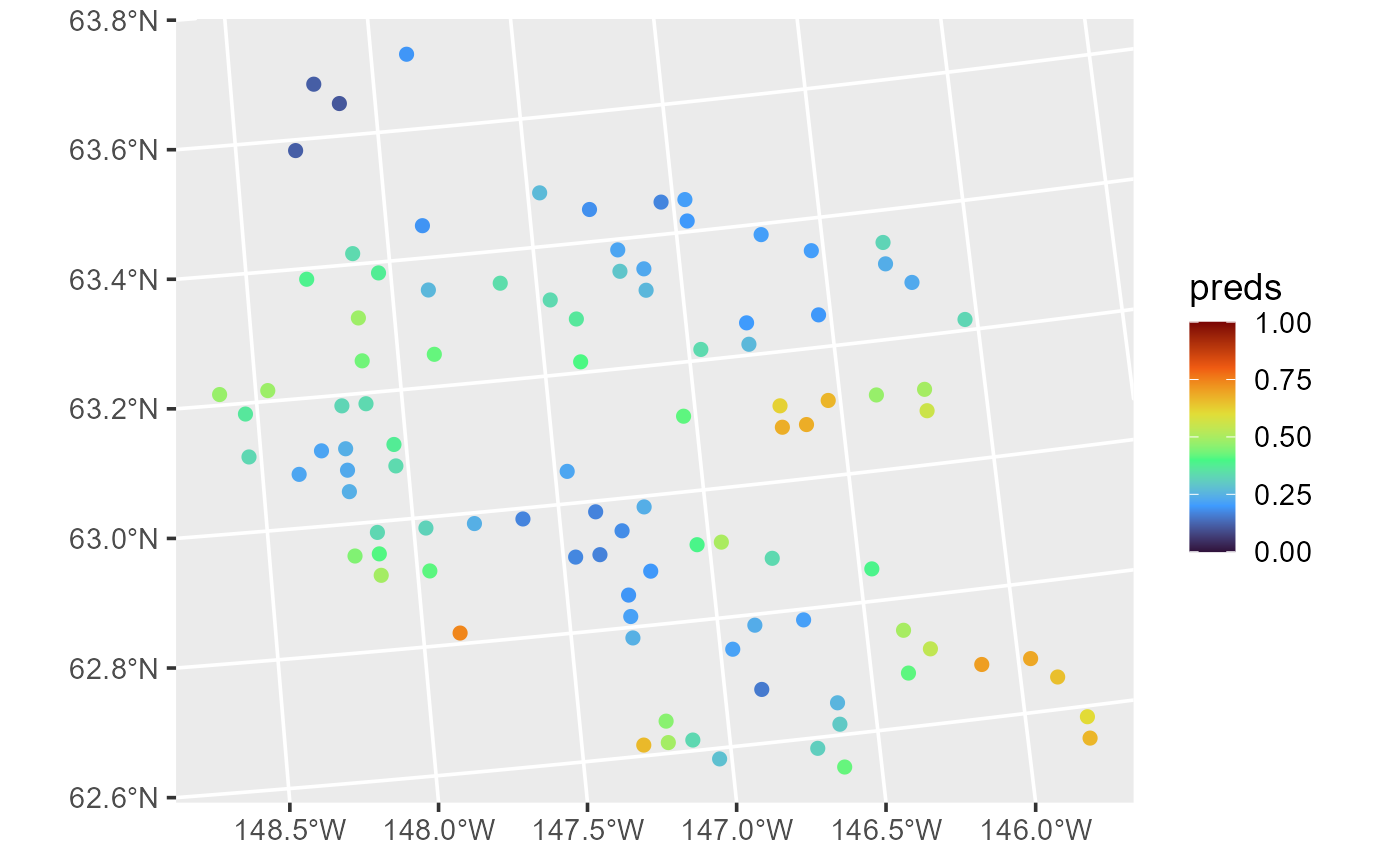
Distribution of moose presence probability predictions in Alaska.
These predictions have similar spatial patterns as moose presence the
observed data. Next we remove the model predictions from
moose_preds and show how augment() can be used
to obtain the same predictions alongside prediction intervals (on the
response scale):
moose_preds$preds <- NULL
augment(binmod, newdata = moose_preds, type.predict = "response", interval = "prediction")#> Simple feature collection with 100 features and 5 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: 269386.2 ymin: 1418453 xmax: 419976.2 ymax: 1541763
#> Projected CRS: NAD83 / Alaska Albers
#> # A tibble: 100 × 6
#> elev strat .fitted .lower .upper geometry
#> * <dbl> <chr> <dbl> <dbl> <dbl> <POINT [m]>
#> 1 143. L 0.705 0.248 0.946 (401239.6 1436192)
#> 2 324. L 0.336 0.0373 0.868 (352640.6 1490695)
#> 3 158. L 0.263 0.0321 0.792 (360954.9 1491590)
#> 4 221. M 0.243 0.0360 0.734 (291839.8 1466091)
#> 5 209. M 0.742 0.270 0.957 (310991.9 1441630)
#> 6 218. L 0.191 0.0196 0.736 (304473.8 1512103)
#> 7 127. L 0.179 0.0226 0.673 (339011.1 1459318)
#> 8 122. L 0.241 0.0344 0.738 (342827.3 1463452)
#> 9 191 L 0.386 0.0414 0.902 (284453.8 1502837)
#> 10 105. L 0.494 0.114 0.882 (391343.9 1483791)
#> # ℹ 90 more rowsSimulating Spatial Generalized Data
Several functions exist to simulate spatial generalized data:
sprbinom() for binomial random variables;
sprbeta() for beta random variables; sprpois()
for Poisson random variables; sprnbinom() for negative
binomial random variables; sprgamma() for gamma random
variables; and spinvgauss() for inverse Gaussian random
variables. All of these functions share similar syntax with
sprnorm() for simulating spatial Gaussian data and require
a spcov_params() object. The generalized data simulation
functions work by first simulating spatial Gaussian data on the link
scale. Then this realization is used as the mean by which to simulate
the spatial generalized linear model data.
We recreate sim_params and sim_data by
running
sim_params <- spcov_params("exponential", de = 5, ie = 1, range = 0.5)
set.seed(0)
n <- 3000
x <- runif(n)
y <- runif(n)
sim_coords <- tibble::tibble(x, y)We simulate a Bernoulli (binomial with size one) random variable by running
sim_response <- sprbinom(sim_params, data = sim_coords, xcoord = x, ycoord = y)
sim_data <- tibble::tibble(sim_coords, sim_response = factor(sim_response))and visualize it by running
ggplot(sim_data, aes(x = x, y = y, color = sim_response)) +
geom_point(size = 1.5) +
scale_color_viridis_d(option = "H") +
theme_gray(base_size = 14)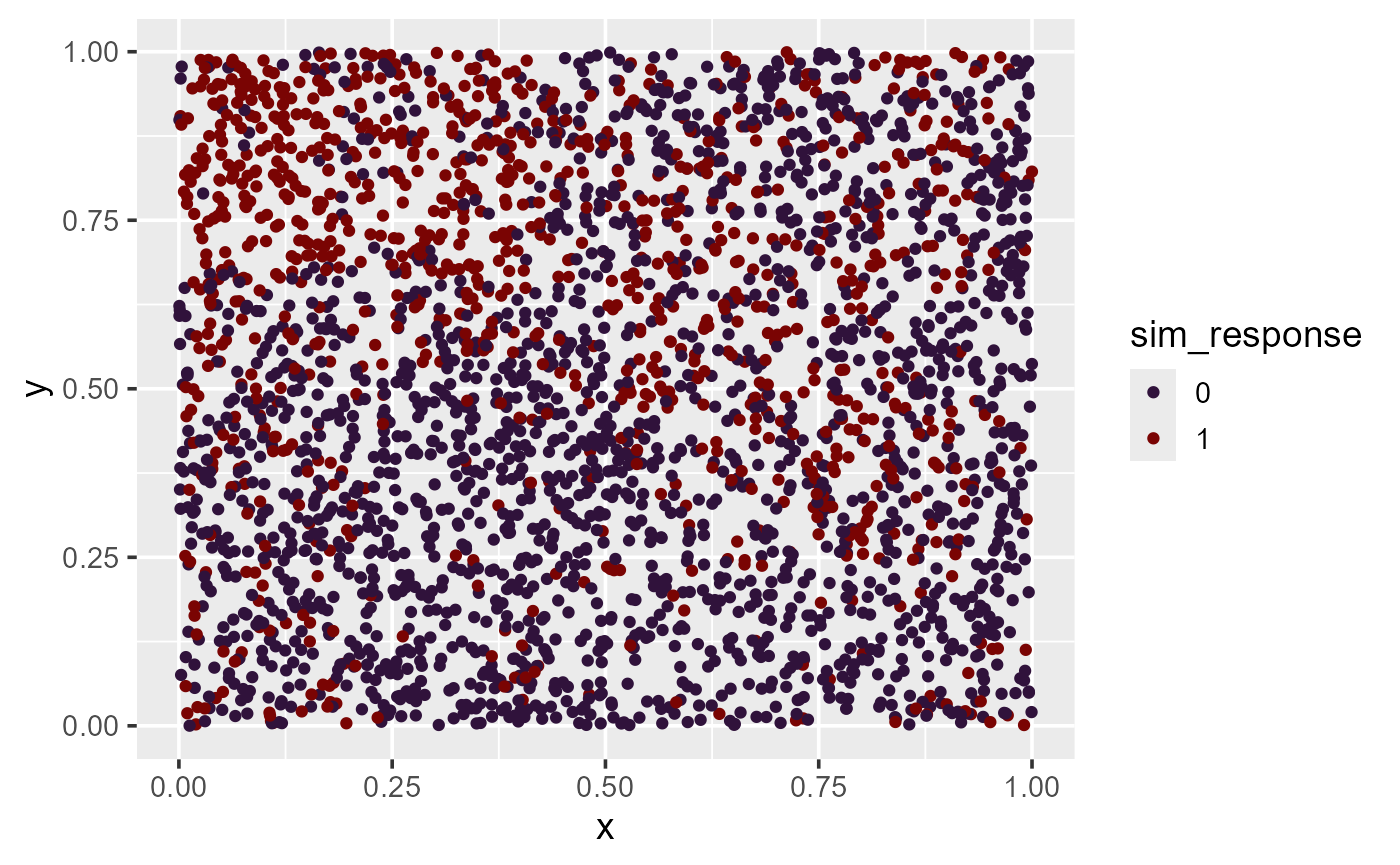
Spatial binomial data simulated in the unit square.
There is noticeable patterning in the response variable
(sim_response) in the same places where there is noticeable
patterning for the spatial Gaussian data. This is expected, as the
spatial Gaussian data are used to inform the mean (proportion) parameter
of the binomial data. Thus where the mean (proportion) parameter is
higher, the more likely the binomial observation takes on the value one
(successes are usually coded as a one).
Additional Examples
We previously showed how to use spmodel to model binary
data via spatial binomial regression. Recall that spmodel
can also be used to model count data via spatial Poisson regression or
spatial negative binomial regression, proportion data via spatial beta
regression, and skewed, positive data via spatial gamma regression or
spatial inverse Gaussian regression. Next we provide examples of fitting
a few of these models.
Poisson regression is used to model count data. To quantify the
relationship between moose counts (count) and elevation, we
fit a spatial Poisson regression model and make predictions by
running
poismod <- spglm(count ~ elev, family = "poisson",
data = moose, spcov_type = "exponential")
predict(poismod, newdata = moose_preds)Poisson regression assumes the mean and variance are equal. Often in practice, the variance is larger than the mean. When the variance is larger than the mean, there is said to be overdispersion in the data. Negative binomial regression accounts for overdispersion via an extra dispersion parameter that allows the variance to change separately from the mean (recall the full negative binomial regression parameterization is available in the technical details vignette). We fit a spatial negative binomial regression model to the moose count data and make predictions by running
nbmod <- spglm(count ~ elev, family = "nbinomial",
data = moose, spcov_type = "exponential")
predict(nbmod, newdata = moose_preds)Gamma regression is used to model skewed, positive data. Suppose
instead of modeling the sulfate data using a spatial linear
model, we modeled sulfate using spatial gamma regression. We fit a
spatial gamma regression model to the sulfate data and make predictions
by running
Discussion
Throughout this vignette, we have shown how to use
spmodel to fit, summarize, and predict for a variety of
spatial statistical models. Spatial linear models for point-referenced
data (i.e., geostatistical models) are fit using the splm()
function while spatial linear models for areal data (i.e., spatial
autoregressive models) are fit using the spautor()
function. Spatial generalized linear models for point-referenced data
(i.e., generalized geostatistical models) are fit using the
spglm() function while spatial generalized linear models
for areal data (i.e., spatial generalized autoregressive models) are fit
using the spgautor() function. Several model-fit statistics
and diagnostics are available. The broom functions tidy()
and glance() are used to tidy and glance at a fitted model.
The broom function augment() is used to augment
data with model diagnostics and augment
newdata with predictions. Several advanced features are
available to accommodate fixed covariance parameter values, random
effects, partition factors, anisotropy, simulated data, big data
approximations, and random forests for model fitting and prediction.
We appreciate feedback from users regarding spmodel. To
learn more about how to provide feedback or contribute to
spmodel, please visit our GitHub repository at https://github.com/USEPA/spmodel.
References
Appendices
An Additional Example Using caribou
The purpose of this example is to show more applications of the
splm() and spautor() functions when the data
are not an sf object, as well as to show a few other
functions in spmodel. The caribou data are are
designed experiment with two treatments observed on an equally spaced
grid and can be analyzed as point-referenced or areal data. We view the
first few rows of caribou by running
caribou#> # A tibble: 30 × 5
#> water tarp z x y
#> <fct> <fct> <dbl> <dbl> <dbl>
#> 1 Y clear 2.42 1 6
#> 2 Y shade 2.44 2 6
#> 3 Y none 1.81 3 6
#> 4 N clear 1.97 4 6
#> 5 N shade 2.38 5 6
#> 6 Y none 2.22 1 5
#> 7 N clear 2.10 2 5
#> 8 Y clear 1.80 3 5
#> 9 Y shade 1.96 4 5
#> 10 Y none 2.10 5 5
#> # ℹ 20 more rowsFirst we analyze caribou as point-referenced data.
Because caribou is not an sf object, we must
provide the columns in caribou that represent the
x-coordinates and y-coordinates. We fit a spatial linear model
regressing nitrogen percentage (z) on water presence
(water) and tarp cover (tarp) by running
cariboumod <- splm(z ~ water + tarp, data = caribou,
spcov_type = "exponential", xcoord = x, ycoord = y)An analysis of variance can be conducted to assess the overall impact
of the tarp variable, which has three levels (clear, shade,
and none), and the water variable, which has two levels
(water and no water). We perform an analysis of variance and tidy the
results by running
#> # A tibble: 3 × 4
#> effects df statistic p.value
#> <chr> <int> <dbl> <dbl>
#> 1 (Intercept) 1 43.5 4.33e-11
#> 2 water 1 1.66 1.98e- 1
#> 3 tarp 2 15.4 4.51e- 4There is significant evidence that at least one tarp cover impacts
nitrogen. Note that, like in summary(), these p-values are
associated with an asymptotic hypothesis test (here, an asymptotic
Chi-squared test).
Next we analyze caribou as areal data. Because
caribou is not an sf object, we must create a
weights matrix. We define two observations as neighbors if they are
adjacent (directly east, west, north, or south) to one another. Two
observations in caribou are adjacent if the distance
between them equals one (recall that observations are not neighbors with
themselves):
Currently, W is a logical matrix with TRUEs
and FALSEs. We coerce it to a numeric matrix by running
W <- W * 1The \(ij\)th value in W
is 1 if the observation in the \(i\)th row is neighbors with the observation
in the \(j\)th row and 0
otherwise. We fit a spatial autoregressive model regressing the nitrogen
percentage (z) on water presence (water) and
tarp cover (tarp) by running
cariboumod <- spautor(z ~ water + tarp, data = caribou,
spcov_type = "car", W = W)We perform an analysis of variance and tidy the results by running
#> # A tibble: 3 × 4
#> effects df statistic p.value
#> <chr> <int> <dbl> <dbl>
#> 1 (Intercept) 1 720. 1.67e-158
#> 2 water 1 1.80 1.80e- 1
#> 3 tarp 2 15.1 5.31e- 4There is significant evidence that at least one tarp cover impacts
nitrogen. Note that, like in summary(), these p-values are
associated with an asymptotic hypothesis test (here, an asymptotic
Chi-squared test).
Random Effect Syntax
A couple of common ways to specify random effects in the
random argument to splm() or
spautor() include:
-
~ (1 | group): Random intercepts for each level ofgroup.~ groupis shorthand for~ (1 | group). -
~ (var | group): Random intercepts for each level ofgroupand random slopes that depend on the variablevarfor each level ofgroup.
Some additional syntax for more complicated random effects structures include:
-
~ (var - 1 | group): Random slopes (without intercepts) that depend on the variablevarfor each level ofgroup. -
~ (1 | group:subgroup): Random intercepts for each combination of levels ingroupand levels insubgroup.~ group:subgroupis shorthand for~ (1 | group:subgroup). -
~ (var | group:subgroup): Random intercepts for each combination of levels ingroupand levels insubgroupand random slopes that depend on the variablevarfor each combination of levels ingroupand levels insubgroup. -
~ (var - 1 | group:subgroup): Random slopes (without intercepts) that depend on the variablevarfor each combination of levels ingroupand levels insubgroup. -
~ (1 | group/subgroup): Shorthand for~ (1 | group) + (1 | group:subgroup). Commonly, thegroup/subgroupnotation impliessubgroupis nested withingroup. -
~ (var | group/subgroup): Shorthand for~ (var | group) + (var | group:subgroup). Commonly, thegroup/subgroupnotation impliessubgroupis nested withingroup. -
~ (var - 1 | group/subgroup): Shorthand for~ (var - 1 | group) + (var - 1 | group:subgroup). Commonly, thegroup/subgroupnotation impliessubgroupis nested withingroup.
Distinct random effects terms are separated in random by
+. Each term must be wrapped in parentheses. For example,
to incorporate random intercepts for group and
subgroup, random looks like
~ (1 | group) + (1 | subgroup). For random intercepts,
recall that ~ group is shorthand for
~ (1 | group). Thus, an equivalent representation of
~ (1 | group) + (1 | subgroup) is
~ group + subgroup. Note that for both random intercepts
and random slopes, the variable on the right-hand side of |
(i.e., group, subgroup,
group:subgroup) must be a factor (or character)
variable.