An Introduction to spmodel
Michael Dumelle, Matt Higham, and Jay M. Ver Hoef
Source:vignettes/introduction.Rmd
introduction.RmdIntroduction
The spmodel R package is used to fit
and summarize spatial models and make predictions at unobserved
locations (Kriging). This vignette provides an overview of basic
features in spmodel. We load spmodel by
running
If you use spmodel in a formal publication or report,
please cite it. Citing spmodel lets us devote more
resources to it in the future. We view the spmodel citation
by running
citation(package = "spmodel")#> To cite spmodel in publications use:
#>
#> Dumelle M, Higham M, Ver Hoef JM (2023). spmodel: Spatial statistical
#> modeling and prediction in R. PLOS ONE 18(3): e0282524.
#> https://doi.org/10.1371/journal.pone.0282524
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Article{,
#> title = {{spmodel}: Spatial statistical modeling and prediction in {R}},
#> author = {Michael Dumelle and Matt Higham and Jay M. {Ver Hoef}},
#> journal = {PLOS ONE},
#> year = {2023},
#> volume = {18},
#> number = {3},
#> pages = {1--32},
#> doi = {10.1371/journal.pone.0282524},
#> url = {https://doi.org/10.1371/journal.pone.0282524},
#> }There are five more spmodel vignettes available on our
website at https://usepa.github.io/spmodel/:
- A Detailed Guide to
spmodel - Spatial Generalized Linear Models in
spmodel - Using
emmeansto Estimate Marginal Means ofspmodelObjects - Block Prediction (i.e., Blocking Kriging) in
spmodel - Technical Details
Additionally, there are two workbooks that have accompanied recent
spmodel workshops:
- 2024 Society for Freshwater Science Conference: “Spatial Analysis
and Statistical Modeling with R and
spmodelavailable at https://usepa.github.io/spworkshop.sfs24/ - 2023 Spatial Statistics Conference:
spmodelworkshop available at https://usepa.github.io/spmodel.spatialstat2023/
The Data
Many of the data sets we use in this vignette are sf
objects. sf objects are data frames (or tibbles) with a
special structure that stores spatial information. They are built using
the sf (Pebesma 2018)
package, which is installed alongside spmodel. We will use
six data sets throughout this vignette:
-
moss: Ansfobject with heavy metal concentrations in Alaska. -
sulfate: Ansfobject with sulfate measurements in the conterminous United States. -
sulfate_preds: Ansfobject with locations at which to predict sulfate measurements in the conterminous United States. -
caribou: Atibble(a specialdata.frame) for a caribou foraging experiment in Alaska. -
moose: Ansfobject with moose measurements in Alaska. -
moose_preds: Ansfobject with locations at which to predict moose measurements in Alaska.
We will create visualizations using ggplot2 (Wickham 2016), which we load by running
ggplot2 is only installed alongside spmodel when
dependencies = TRUE in install.packages(), so
check that it is installed before reproducing any visualizations in this
vignette.
Spatial Linear Models
Spatial linear models for a quantitative response vector \(\mathbf{y}\) have spatially dependent random errors and are often parameterized as
\[ \mathbf{y} = \mathbf{X} \boldsymbol{\beta} + \boldsymbol{\tau} + \boldsymbol{\epsilon}, \]
where \(\mathbf{X}\) is a matrix of explanatory variables (usually including a column of 1’s for an intercept), \(\boldsymbol{\beta}\) is a vector of fixed effects that describe the average impact of \(\mathbf{X}\) on \(\mathbf{y}\), \(\boldsymbol{\tau}\) is a vector of spatially dependent (correlated) random errors, and \(\boldsymbol{\epsilon}\) is a vector of spatially independent (uncorrelated) random errors. The spatial dependence of \(\boldsymbol{\tau}\) is explicitly specified using a spatial covariance function that incorporates the variance of \(\boldsymbol{\tau}\), often called the partial sill, and a range parameter that controls the behavior of the spatial covariance. The variance of \(\boldsymbol{\epsilon}\) is often called the nugget.
Spatial linear models are fit in spmodel for
point-referenced and areal data. Data are point-referenced when the
elements in \(\mathbf{y}\) are observed
at point-locations indexed by x-coordinates and y-coordinates on a
spatially continuous surface with an infinite number of locations. The
splm() function is used to fit spatial linear models for
point-referenced data (these are often called geostatistical models).
Data are areal when the elements in \(\mathbf{y}\) are observed as part of a
finite network of polygons whose connections are indexed by a
neighborhood structure. For example, the polygons may represent counties
in a state who are neighbors if they share at least one boundary. The
spautor() function is used to fit spatial linear models for
areal data (these are often called spatial autoregressive models). This
vignette focuses on spatial linear models for point-referenced data,
though spmodel’s other vignettes discuss spatial linear
models for areal data.
The splm() function has similar syntax and output as the
commonly used lm() function used to fit non-spatial linear
models. splm() generally requires at least three
arguments:
-
formula: a formula that describes the relationship between the response variable and explanatory variables.-
formulauses the same syntax as theformulaargument inlm()
-
-
data: adata.frameorsfobject that contains the response variable, explanatory variables, and spatial information. -
spcov_type: the spatial covariance type ("exponential","spherical","matern", etc).
If data is an sf object, the coordinate
information is taken from the object’s geometry. If data is
a data.frame (or tibble), then
xcoord and ycoord are required arguments to
splm() that specify the columns in data
representing the x-coordinates and y-coordinates, respectively.
spmodel uses the spatial coordinates “as-is,” meaning that
spmodel does not perform any projections. To project your
data or change the coordinate reference system, use
sf::st_transform(). If an sf object with
polygon geometries is given to splm(), the centroids of
each polygon are used to fit the spatial linear model.
Next we show the basic features and syntax of splm()
using the Alaskan moss data. We study the impact of log
distance to the road (log_dist2road) on log zinc
concentration (log_Zn). We view the first few rows of the
moss data by running
moss#> Simple feature collection with 365 features and 7 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: -445884.1 ymin: 1929616 xmax: -383656.8 ymax: 2061414
#> Projected CRS: NAD83 / Alaska Albers
#> # A tibble: 365 × 8
#> sample field_dup lab_rep year sideroad log_dist2road log_Zn
#> <fct> <fct> <fct> <fct> <fct> <dbl> <dbl>
#> 1 001PR 1 1 2001 N 2.68 7.33
#> 2 001PR 1 2 2001 N 2.68 7.38
#> 3 002PR 1 1 2001 N 2.54 7.58
#> 4 003PR 1 1 2001 N 2.97 7.63
#> 5 004PR 1 1 2001 N 2.72 7.26
#> 6 005PR 1 1 2001 N 2.76 7.65
#> 7 006PR 1 1 2001 S 2.30 7.59
#> 8 007PR 1 1 2001 N 2.78 7.16
#> 9 008PR 1 1 2001 N 2.93 7.19
#> 10 009PR 1 1 2001 N 2.79 8.07
#> # ℹ 355 more rows
#> # ℹ 1 more variable: geometry <POINT [m]>We can visualize the distribution of log zinc concentration
(log_Zn) by running
ggplot(moss, aes(color = log_Zn)) +
geom_sf() +
scale_color_viridis_c()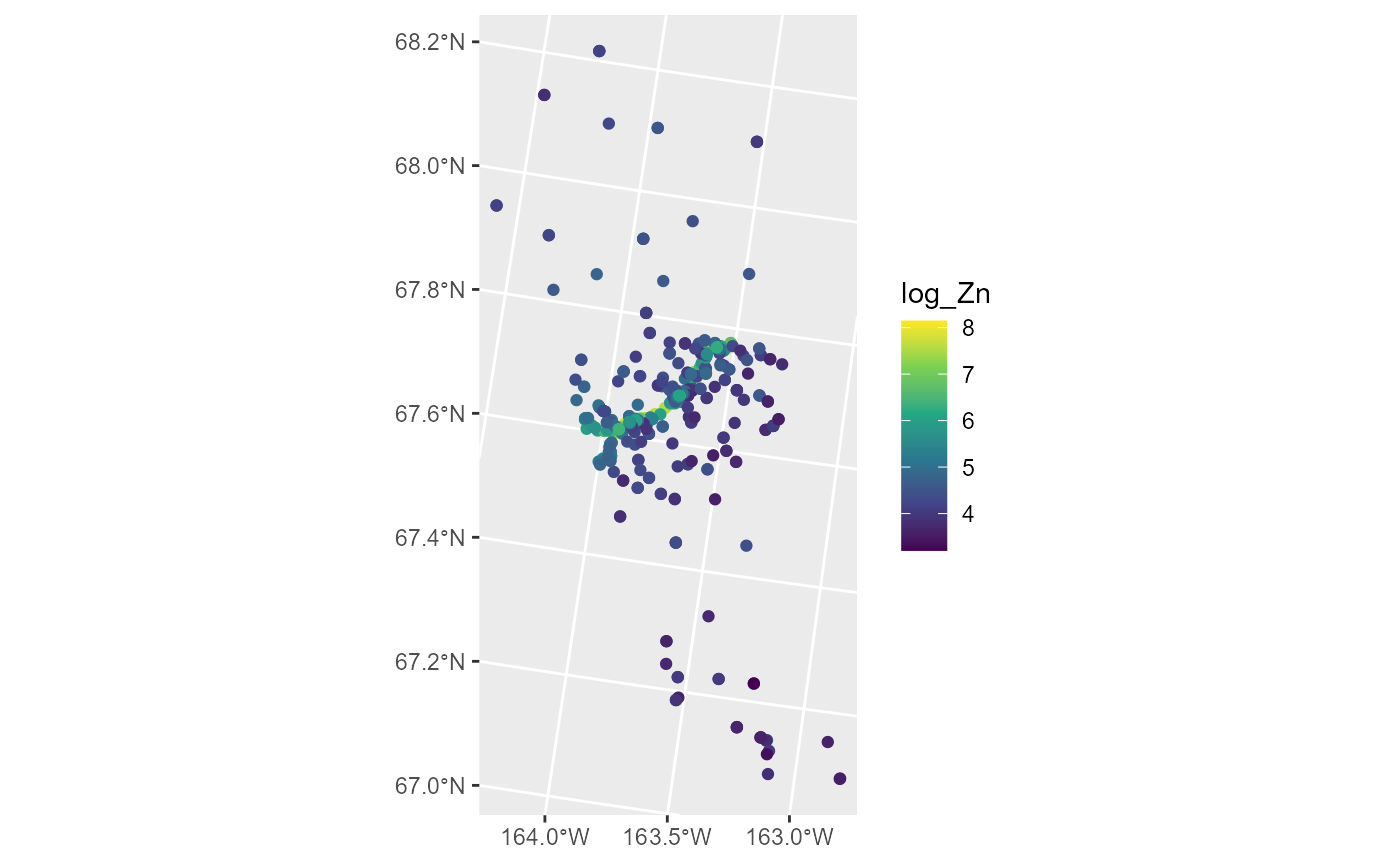
Log zinc concentration appears highest in the middle of the spatial domain, which has a road running through it. We fit a spatial linear model regressing log zinc concentration on log distance to the road using an exponential spatial covariance function by running
spmod <- splm(log_Zn ~ log_dist2road, data = moss, spcov_type = "exponential")The estimation method is specified via the estmethod
argument, which has a default value of "reml" for
restricted maximum likelihood. Other estimation methods are
"ml" for maximum likelihood, "sv-wls" for
semivariogram weighted least squares, and "sv-cl" for
semivariogram composite likelihood.
Printing spmod shows the function call, the estimated
fixed effect coefficients, and the estimated spatial covariance
parameters. de is the estimated variance of \(\boldsymbol{\tau}\) (the spatially
dependent random error), ie is the estimated variance of
\(\boldsymbol{\epsilon}\) (the
spatially independent random error), and range is the range
parameter.
print(spmod)#>
#> Call:
#> splm(formula = log_Zn ~ log_dist2road, data = moss, spcov_type = "exponential")
#>
#>
#> Coefficients (fixed):
#> (Intercept) log_dist2road
#> 9.7683 -0.5629
#>
#>
#> Coefficients (exponential spatial covariance):
#> de ie range
#> 3.595e-01 7.897e-02 8.237e+03Next we show how to obtain more detailed summary information from the fitted model.
Model Summaries
We summarize the fitted model by running
summary(spmod)#>
#> Call:
#> splm(formula = log_Zn ~ log_dist2road, data = moss, spcov_type = "exponential")
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -2.6801 -1.3606 -0.8103 -0.2485 1.1298
#>
#> Coefficients (fixed):
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 9.76825 0.25216 38.74 <2e-16 ***
#> log_dist2road -0.56287 0.02013 -27.96 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Pseudo R-squared: 0.683
#>
#> Coefficients (exponential spatial covariance):
#> de ie range
#> 3.595e-01 7.897e-02 8.237e+03Similar to summaries of lm() objects, summaries of
splm() objects include the original function call,
residuals, and a coefficients table of fixed effects. Log zinc
concentration appears to significantly decrease with log distance from
the road, as evidenced by the small p-value associated with the
asymptotic z-test. A pseudo r-squared is also returned, which quantifies
the proportion of variability explained by the fixed effects.
In the remainder of this subsection, we describe the broom (Robinson, Hayes, and Couch 2021) functions
tidy(), glance() and augment().
tidy() tidies coefficient output in a convenient
tibble, glance() glances at model-fit
statistics, and augment() augments the data with fitted
model diagnostics.
We tidy the fixed effects by running
tidy(spmod)#> # A tibble: 2 × 5
#> term estimate std.error statistic p.value
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 (Intercept) 9.77 0.252 38.7 0
#> 2 log_dist2road -0.563 0.0201 -28.0 0We glance at the model-fit statistics by running
glance(spmod)#> # A tibble: 1 × 10
#> n p npar value AIC AICc BIC logLik deviance pseudo.r.squared
#> <int> <dbl> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 365 2 3 367. 373. 373. 385. -184. 363. 0.683The columns of this tibble represent:
-
n: The sample size -
p: The number of fixed effects (linearly independent columns in \(\mathbf{X}\)) -
npar: The number of estimated covariance parameters -
value: The value of the minimized objective function used when fitting the model -
AIC: The Akaike Information Criterion (AIC) -
AICc: The AIC with a small sample size correction -
BIC: The Bayesian Information Criterion (BIC) -
logLik: The log-likelihood -
deviance: The deviance -
pseudo.r.squared: The pseudo r-squared
The glances() function can be used to glance at multiple
models at once. Suppose we wanted to compare the current model, which
uses an exponential spatial covariance, to a new model without spatial
covariance (equivalent to a model fit using lm()). We do
this using glances() by running
#> # A tibble: 2 × 11
#> model n p npar value AIC AICc BIC logLik deviance
#> <chr> <int> <dbl> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 spmod 365 2 3 367. 373. 373. 385. -184. 363.
#> 2 lmod 365 2 1 632. 634. 634. 638. -316. 363.
#> # ℹ 1 more variable: pseudo.r.squared <dbl>The much lower AIC and AICc for the spatial linear model indicates it
is a much better fit to the data. Outside of glance() and
glances(), the functions AIC(),
AICc(), BIC() logLik(),
deviance(), and pseudoR2() are available to
compute the relevant statistics.
We augment the data with diagnostics by running
augment(spmod)#> Simple feature collection with 365 features and 7 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: -445884.1 ymin: 1929616 xmax: -383656.8 ymax: 2061414
#> Projected CRS: NAD83 / Alaska Albers
#> # A tibble: 365 × 8
#> log_Zn log_dist2road .fitted .resid .hat .cooksd .std.resid
#> * <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 7.33 2.68 8.26 -0.928 0.0200 0.0142 1.18
#> 2 7.38 2.68 8.26 -0.880 0.0200 0.0186 1.35
#> 3 7.58 2.54 8.34 -0.755 0.0225 0.00482 0.647
#> 4 7.63 2.97 8.09 -0.464 0.0197 0.0305 1.74
#> 5 7.26 2.72 8.24 -0.977 0.0215 0.131 3.45
#> 6 7.65 2.76 8.21 -0.568 0.0284 0.0521 1.89
#> 7 7.59 2.30 8.47 -0.886 0.0300 0.0591 1.96
#> 8 7.16 2.78 8.20 -1.05 0.0335 0.00334 0.439
#> 9 7.19 2.93 8.12 -0.926 0.0378 0.0309 1.26
#> 10 8.07 2.79 8.20 -0.123 0.0314 0.00847 0.723
#> # ℹ 355 more rows
#> # ℹ 1 more variable: geometry <POINT [m]>The columns of this tibble represent:
-
log_Zn: The log zinc concentration. -
log_dist2road: The log distance to the road. -
.fitted: The fitted values (the estimated mean given the explanatory variable values). -
.resid: The residuals (the response minus the fitted values). -
.hat: The leverage (hat) values. -
.cooksd: The Cook’s distance -
.std.residuals: Standardized residuals -
geometry: The spatial information in thesfobject.
By default, augment() only returns the variables in the
data used by the model. All variables from the original data are
returned by setting drop = FALSE. Many of these model
diagnostics can be visualized by running plot(spmod). We
can learn more about plot() in spmodel by
running help("plot.spmodel", "spmodel").
Prediction (Kriging)
Commonly a goal of a data analysis is to make predictions at
unobserved locations. In spatial contexts, prediction is often called
Kriging. Next we use the sulfate data to build a spatial
linear model of sulfate measurements in the conterminous United States
with the goal of making sulfate predictions (Kriging) for the unobserved
locations in sulfate_preds.
We visualize the distribution of sulfate by running
ggplot(sulfate, aes(color = sulfate)) +
geom_sf(size = 2) +
scale_color_viridis_c(limits = c(0, 45))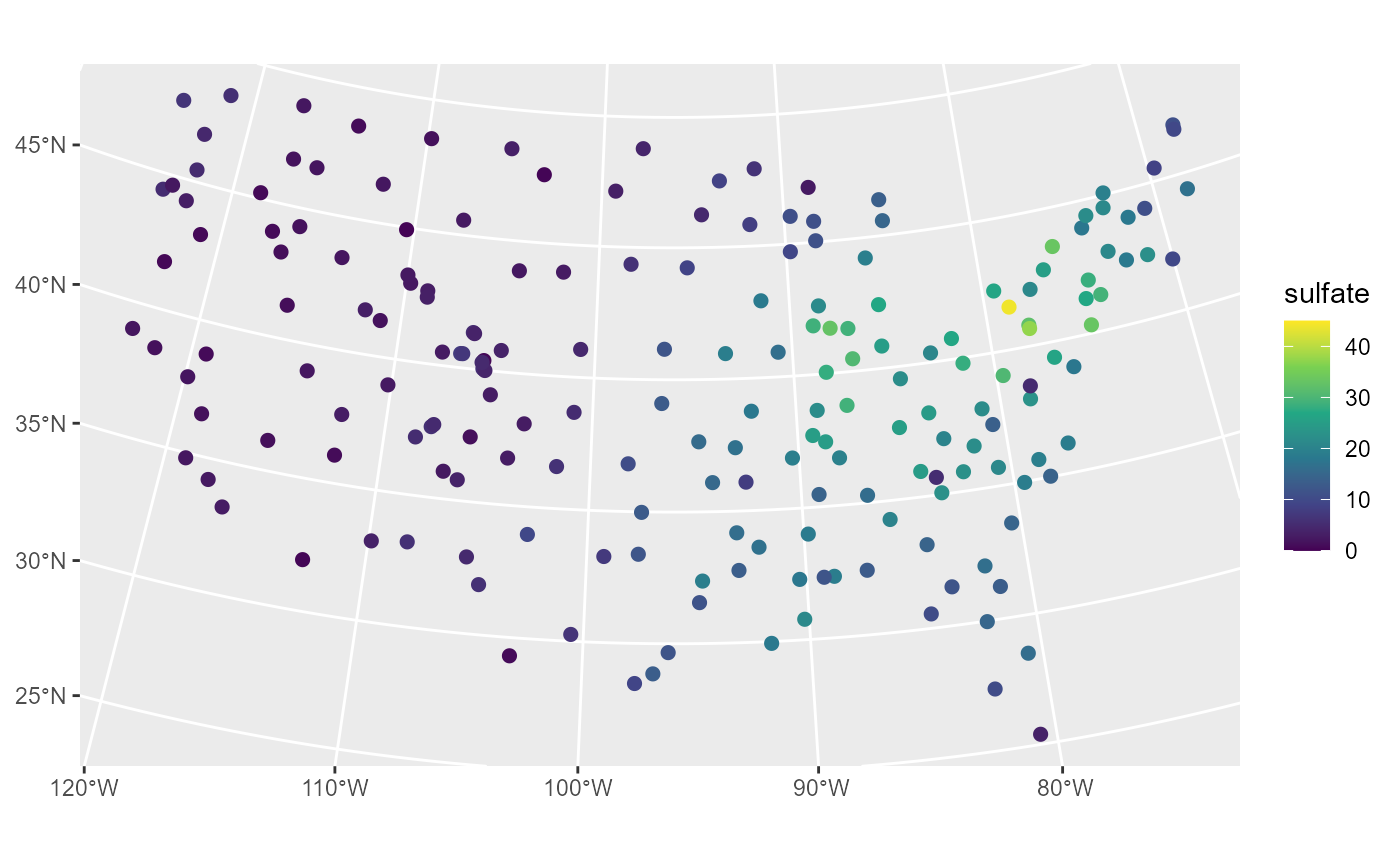
Sulfate appears spatially dependent, as measurements are highest in the Northeast and lowest in the Midwest and West.
We fit a spatial linear model regressing sulfate on an intercept using a spherical spatial covariance function by running
sulfmod <- splm(sulfate ~ 1, data = sulfate, spcov_type = "spherical")We make predictions at the locations in sulfate_preds
and store them as a new variable called preds in the
sulfate_preds data set by running
sulfate_preds$preds <- predict(sulfmod, newdata = sulfate_preds)We visualize these predictions by running
ggplot(sulfate_preds, aes(color = preds)) +
geom_sf(size = 2) +
scale_color_viridis_c(limits = c(0, 45))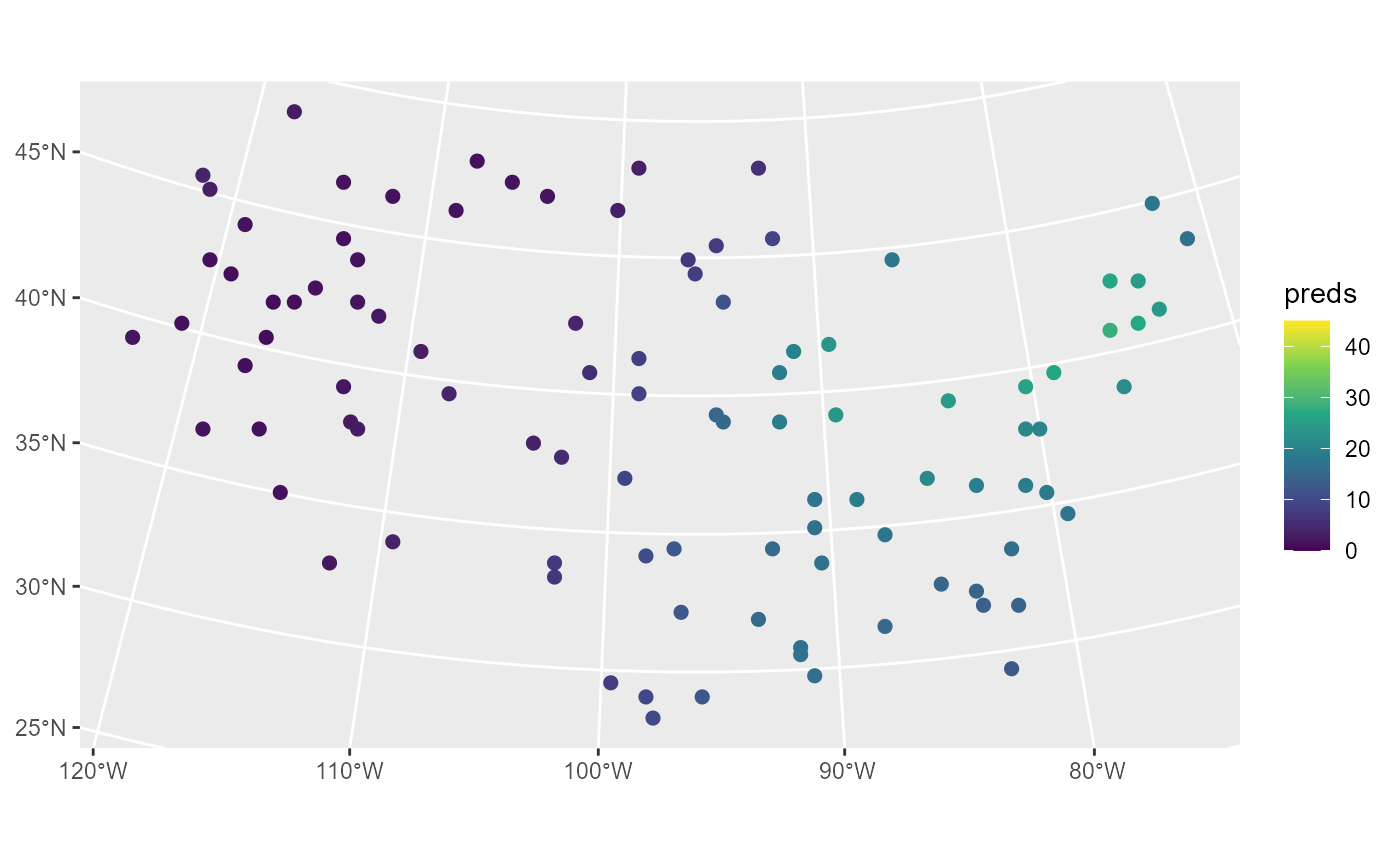
These predictions have similar sulfate patterns as in the observed
data (predicted values are highest in the Northeast and lowest in the
Midwest and West). Next we remove the model predictions from
sulfate_preds before showing how augment() can
be used to obtain the same predictions:
sulfate_preds$preds <- NULLWhile augment() was previously used to augment the
original data with model diagnostics, it can also be used to augment the
newdata data with predictions:
augment(sulfmod, newdata = sulfate_preds)#> Simple feature collection with 100 features and 1 field
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: -2283774 ymin: 582930.5 xmax: 1985906 ymax: 3037173
#> Projected CRS: NAD83 / Conus Albers
#> # A tibble: 100 × 2
#> .fitted geometry
#> * <dbl> <POINT [m]>
#> 1 1.40 (-1771413 1752976)
#> 2 24.5 (1018112 1867127)
#> 3 8.99 (-291256.8 1553212)
#> 4 16.4 (1274293 1267835)
#> 5 4.91 (-547437.6 1638825)
#> 6 26.7 (1445080 1981278)
#> 7 3.00 (-1629090 3037173)
#> 8 14.3 (1302757 1039534)
#> 9 1.49 (-1429838 2523494)
#> 10 14.4 (1131970 1096609)
#> # ℹ 90 more rowsHere .fitted represents the predictions.
Confidence intervals for the mean response or prediction intervals
for the predicted response can be obtained by specifying the
interval argument in predict() and
augment():
augment(sulfmod, newdata = sulfate_preds, interval = "prediction")#> Simple feature collection with 100 features and 3 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: -2283774 ymin: 582930.5 xmax: 1985906 ymax: 3037173
#> Projected CRS: NAD83 / Conus Albers
#> # A tibble: 100 × 4
#> .fitted .lower .upper geometry
#> * <dbl> <dbl> <dbl> <POINT [m]>
#> 1 1.40 -6.62 9.42 (-1771413 1752976)
#> 2 24.5 17.0 32.0 (1018112 1867127)
#> 3 8.99 1.09 16.9 (-291256.8 1553212)
#> 4 16.4 8.67 24.2 (1274293 1267835)
#> 5 4.91 -2.80 12.6 (-547437.6 1638825)
#> 6 26.7 19.2 34.2 (1445080 1981278)
#> 7 3.00 -4.92 10.9 (-1629090 3037173)
#> 8 14.3 6.76 21.8 (1302757 1039534)
#> 9 1.49 -6.34 9.32 (-1429838 2523494)
#> 10 14.4 6.74 22.1 (1131970 1096609)
#> # ℹ 90 more rowsBy default, predict() and augment() compute
95% intervals, though this can be changed using the level
argument.
While the fitted model in this example only used an intercept, the
same code is used for prediction with fitted models having explanatory
variables. If explanatory variables were used to fit the model, the same
explanatory variables must be included in newdata with the
same names they have in data. If data is a
data.frame, coordinates must be included in
newdata with the same names as they have in
data. If data is an sf object,
coordinates must be included in newdata with the same
geometry name as they have in data. When using projected
coordinates, the projection for newdata should be the same
as the projection for data.
An Additional Example
We now use the caribou data from a foraging experiment
conducted in Alaska to show an application of splm() to
data stored in a tibble (data.frame) instead
of an sf object. In caribou, the x-coordinates
are stored in the x column and the y-coordinates are stored
in the y column. We view the first few rows of
caribou by running
caribou#> # A tibble: 30 × 5
#> water tarp z x y
#> <fct> <fct> <dbl> <dbl> <dbl>
#> 1 Y clear 2.42 1 6
#> 2 Y shade 2.44 2 6
#> 3 Y none 1.81 3 6
#> 4 N clear 1.97 4 6
#> 5 N shade 2.38 5 6
#> 6 Y none 2.22 1 5
#> 7 N clear 2.10 2 5
#> 8 Y clear 1.80 3 5
#> 9 Y shade 1.96 4 5
#> 10 Y none 2.10 5 5
#> # ℹ 20 more rowsWe fit a spatial linear model regressing nitrogen percentage
(z) on water presence (water) and tarp cover
(tarp) by running
cariboumod <- splm(z ~ water + tarp, data = caribou,
spcov_type = "exponential", xcoord = x, ycoord = y)An analysis of variance can be conducted to assess the overall impact
of the tarp variable, which has three levels (clear, shade,
and none), and the water variable, which has two levels
(water and no water). We perform an analysis of variance by running
anova(cariboumod)#> Analysis of Variance Table
#>
#> Response: z
#> Df Chi2 Pr(>Chi2)
#> (Intercept) 1 43.4600 4.327e-11 ***
#> water 1 1.6603 0.1975631
#> tarp 2 15.4071 0.0004512 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1There seems to be significant evidence that at least one tarp cover
impacts nitrogen. Note that, like in summary(), these
p-values are associated with an asymptotic hypothesis test (here, an
asymptotic Chi-squared test).
Spatial Generalized Linear Models
When building spatial linear models, the response vector \(\mathbf{y}\) is typically assumed Gaussian
(given \(\mathbf{X}\)). Relaxing this
assumption on the distribution of \(\mathbf{y}\) yields a rich class of spatial
generalized linear models that can describe binary data, proportion
data, count data, and skewed data. Spatial generalized linear models are
parameterized as \[
g(\boldsymbol{\mu}) = \boldsymbol{\eta} = \mathbf{X} \boldsymbol{\beta}
+ \boldsymbol{\tau} + \boldsymbol{\epsilon},
\] where \(g(\cdot)\) is called
a link function, \(\boldsymbol{\mu}\)
is the mean of \(\mathbf{y}\), and the
remaining terms \(\mathbf{X}\), \(\boldsymbol{\beta}\), \(\boldsymbol{\tau}\), \(\boldsymbol{\epsilon}\) represent the same
quantities as for the spatial linear models. The link function, \(g(\cdot)\), “links” a function of \(\boldsymbol{\mu}\) to the linear term \(\boldsymbol{\eta}\), denoted here as \(\mathbf{X} \boldsymbol{\beta} + \boldsymbol{\tau}
+ \boldsymbol{\epsilon}\), which is familiar from spatial linear
models. Note that the linking of \(\boldsymbol{\mu}\) to \(\boldsymbol{\eta}\) applies element-wise to
each vector. Each link function \(g(\cdot)\) has a corresponding inverse link
function, \(g^{-1}(\cdot)\). The
inverse link function “links” a function of \(\boldsymbol{\eta}\) to \(\boldsymbol{\mu}\). Notice that for spatial
generalized linear models, we are not modeling \(\mathbf{y}\) directly as we do for spatial
linear models, but rather we are modeling a function of the mean of
\(\mathbf{y}\). Also notice that \(\boldsymbol{\eta}\) is unconstrained but
\(\boldsymbol{\mu}\) is usually
constrained in some way (e.g., positive). Next we discuss the specific
distributions and link functions used in spmodel.
spmodel allows fitting of spatial generalized linear
models when \(\mathbf{y}\) is a
binomial (or Bernoulli), beta, Poisson, negative binomial, gamma, or
inverse Gaussian random vector. For binomial and beta \(\mathbf{y}\), the logit link function is
defined as \(g(\boldsymbol{\mu}) =
\ln(\frac{\boldsymbol{\mu}}{1 - \boldsymbol{\mu}}) =
\boldsymbol{\eta}\), and the inverse logit link function is
defined as \(g^{-1}(\boldsymbol{\eta}) =
\frac{\exp(\boldsymbol{\eta})}{1 + \exp(\boldsymbol{\eta})} =
\boldsymbol{\mu}\). For Poisson, negative binomial, gamma, and
inverse Gaussian \(\mathbf{y}\), the
log link function is defined as \(g(\boldsymbol{\mu}) = \ln(\boldsymbol{\mu}) =
\boldsymbol{\eta}\), and the inverse log link function is defined
as \(g^{-1}(\boldsymbol{\eta}) =
\exp(\boldsymbol{\eta}) = \boldsymbol{\mu}\).
As with spatial linear models, spatial generalized linear models are
fit in spmodel for point-referenced and areal data. The
spglm() function is used to fit spatial generalized linear
models for point-referenced data, and the spgautor()
function is used to fit spatial generalized linear models for areal
data. Though this vignette focuses on point-referenced data,
spmodel’s other vignettes discuss spatial generalized
linear models for areal data.
The spglm() function is quite similar to the
splm() function, though one additional argument is
required:
-
family: the generalized linear model family (i.e., the distribution of \(\mathbf{y}\)).familycan bebinomial,beta,poisson,nbinomial,Gamma, orinverse.gaussian.
Next we show the basic features and syntax of spglm()
using the moose data. We study the impact of elevation
(elev) on the presence of moose (presence)
observed at a site location in Alaska. presence equals one
if at least one moose was observed at the site and zero otherwise. We
view the first few rows of the moose data by running
moose#> Simple feature collection with 218 features and 4 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: 269085 ymin: 1416151 xmax: 419976.2 ymax: 1541763
#> Projected CRS: NAD83 / Alaska Albers
#> # A tibble: 218 × 5
#> elev strat count presence geometry
#> <dbl> <chr> <dbl> <fct> <POINT [m]>
#> 1 469. L 0 0 (293542.6 1541016)
#> 2 362. L 0 0 (298313.1 1533972)
#> 3 173. M 0 0 (281896.4 1532516)
#> 4 280. L 0 0 (298651.3 1530264)
#> 5 620. L 0 0 (311325.3 1527705)
#> 6 164. M 0 0 (291421.5 1518398)
#> 7 164. M 0 0 (287298.3 1518035)
#> 8 186. L 0 0 (279050.9 1517324)
#> 9 362. L 0 0 (346145.9 1512479)
#> 10 430. L 0 0 (321354.6 1509966)
#> # ℹ 208 more rowsWe can visualize the distribution of moose presence by running
ggplot(moose, aes(color = presence)) +
scale_color_viridis_d(option = "H") +
geom_sf(size = 2) 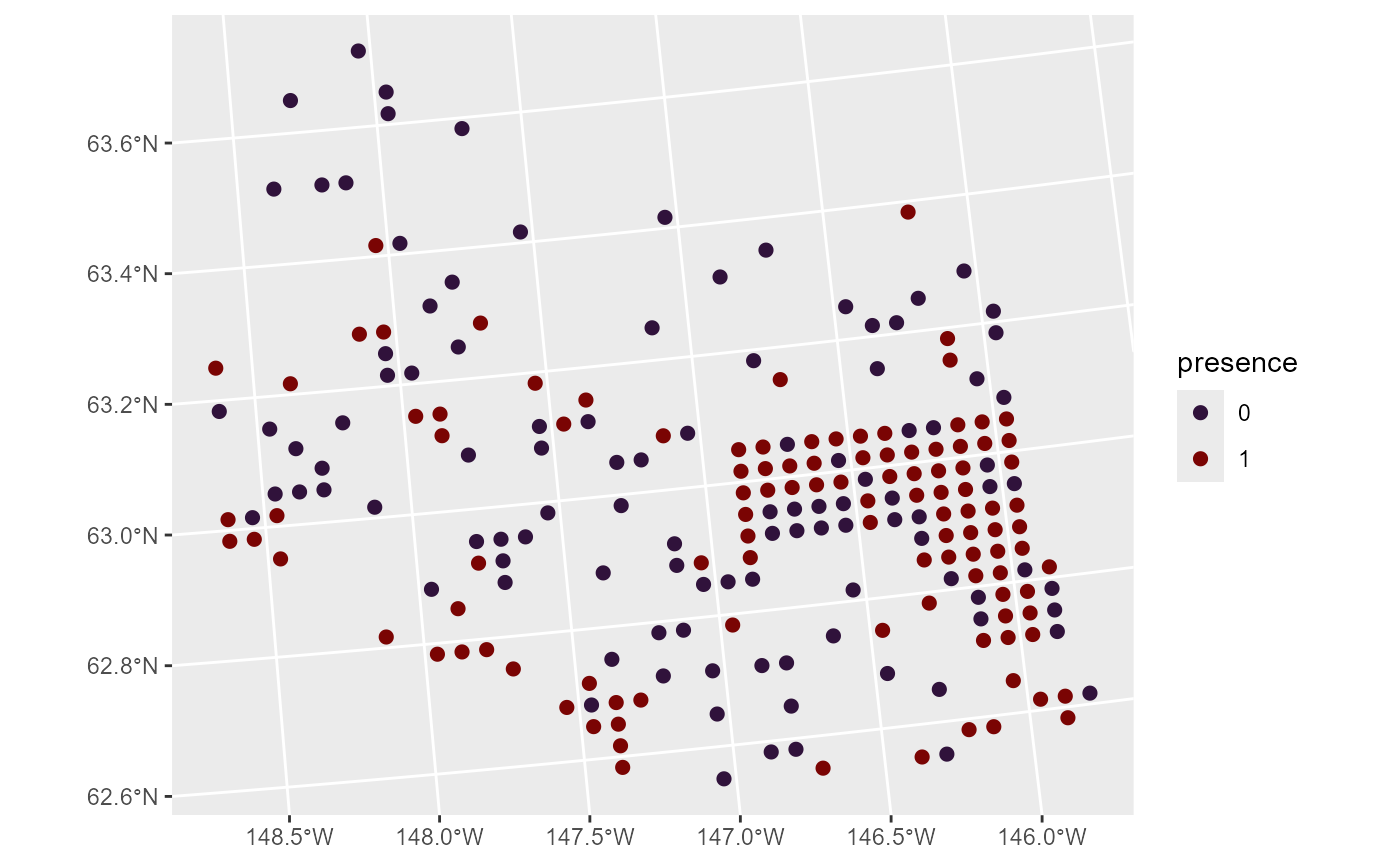
One example of a generalized linear model is a binomial (e.g., logistic) regression model. Binomial regression models are often used to model presence data such as this. To quantify the relationship between moose presence and elevation, we fit a spatial binomial regression model (a specific spatial generalized linear model) by running
binmod <- spglm(presence ~ elev, family = "binomial",
data = moose, spcov_type = "exponential")The estimation method is specified via the estmethod
argument, which has a default value of "reml" for
restricted maximum likelihood. The other estimation method is
"ml" for maximum likelihood.
Printing binmod shows the function call, the estimated
fixed effect coefficients (on the link scale), the estimated spatial
covariance parameters, and a dispersion parameter. The dispersion
parameter is estimated for some spatial generalized linear models and
changes the mean-variance relationship of \(\mathbf{y}\). For binomial regression
models, the dispersion parameter is not estimated and is always fixed at
one.
print(binmod)#>
#> Call:
#> spglm(formula = presence ~ elev, family = "binomial", data = moose,
#> spcov_type = "exponential")
#>
#>
#> Coefficients (fixed):
#> (Intercept) elev
#> -0.874038 0.002365
#>
#>
#> Coefficients (exponential spatial covariance):
#> de ie range
#> 3.746e+00 4.392e-03 3.203e+04
#>
#>
#> Coefficients (Dispersion for binomial family):
#> dispersion
#> 1Model Summaries
We summarize the fitted model by running
summary(binmod)#>
#> Call:
#> spglm(formula = presence ~ elev, family = "binomial", data = moose,
#> spcov_type = "exponential")
#>
#> Deviance Residuals:
#> Min 1Q Median 3Q Max
#> -1.5249 -0.8114 0.5600 0.8306 1.5757
#>
#> Coefficients (fixed):
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -0.874038 1.140953 -0.766 0.444
#> elev 0.002365 0.003184 0.743 0.458
#>
#> Pseudo R-squared: 0.00311
#>
#> Coefficients (exponential spatial covariance):
#> de ie range
#> 3.746e+00 4.392e-03 3.203e+04
#>
#> Coefficients (Dispersion for binomial family):
#> dispersion
#> 1Similar to summaries of glm() objects, summaries of
spglm() objects include the original function call, summary
statistics of the deviance residuals, and a coefficients table of fixed
effects. The logit of moose presence probability does not appear to be
related to elevation, as evidenced by the large p-value associated with
the asymptotic z-test. A pseudo r-squared is also returned, which
quantifies the proportion of variability explained by the fixed effects.
The spatial covariance parameters and dispersion parameter are also
returned.
The tidy(), glance(), and
augment() functions behave similarly for
spglm() objects as they do for splm() objects.
We tidy the fixed effects (on the link scale) by running
tidy(binmod)#> # A tibble: 2 × 5
#> term estimate std.error statistic p.value
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 (Intercept) -0.874 1.14 -0.766 0.444
#> 2 elev 0.00237 0.00318 0.743 0.458We glance at the model-fit statistics by running
glance(binmod)#> # A tibble: 1 × 10
#> n p npar value AIC AICc BIC logLik deviance pseudo.r.squared
#> <int> <dbl> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 218 2 3 692. 698. 698. 708. -346. 190. 0.00311We glance at the spatial binomial regression model and a non-spatial binomial regression model by running
glmod <- spglm(presence ~ elev, family = "binomial", data = moose, spcov_type = "none")
glances(binmod, glmod)#> # A tibble: 2 × 11
#> model n p npar value AIC AICc BIC logLik deviance
#> <chr> <int> <dbl> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 binmod 218 2 3 692. 698. 698. 708. -346. 190.
#> 2 glmod 218 2 0 715. 715. 715. 715. -358. 302.
#> # ℹ 1 more variable: pseudo.r.squared <dbl>The lower AIC and AICc for the spatial binomial regression model indicates it is a much better fit to the data.
We augment the data with diagnostics by running
augment(binmod)#> Simple feature collection with 218 features and 7 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: 269085 ymin: 1416151 xmax: 419057.4 ymax: 1541016
#> Projected CRS: NAD83 / Alaska Albers
#> # A tibble: 218 × 8
#> presence elev .fitted .resid .hat .cooksd .std.resid
#> * <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 0 469. -1.73 -0.571 0.0500 0.00904 -0.586
#> 2 0 362. -2.12 -0.477 0.0168 0.00198 -0.481
#> 3 0 173. -2.15 -0.468 0.00213 0.000235 -0.469
#> 4 0 280. -2.27 -0.444 0.00616 0.000615 -0.445
#> 5 0 620. -1.40 -0.664 0.136 0.0402 -0.714
#> 6 0 164. -1.90 -0.528 0.00260 0.000364 -0.528
#> 7 0 164. -1.86 -0.538 0.00269 0.000392 -0.539
#> 8 0 186. -1.61 -0.603 0.00332 0.000607 -0.604
#> 9 0 362. -1.60 -0.606 0.0245 0.00474 -0.614
#> 10 0 430. -1.24 -0.714 0.0528 0.0150 -0.734
#> # ℹ 208 more rows
#> # ℹ 1 more variable: geometry <POINT [m]>Prediction (Kriging)
For spatial generalized linear models, we are predicting the mean of
the process generating the observation rather than the observation
itself. We make predictions of moose presence probability at the
locations in moose_preds by running
moose_preds$preds <- predict(binmod, newdata = moose_preds, type = "response")The type argument specifies whether predictions are returned on the link or response (inverse link) scale. We visualize these predictions by running
ggplot(moose_preds, aes(color = preds)) +
geom_sf(size = 2) +
scale_color_viridis_c(limits = c(0, 1), option = "H")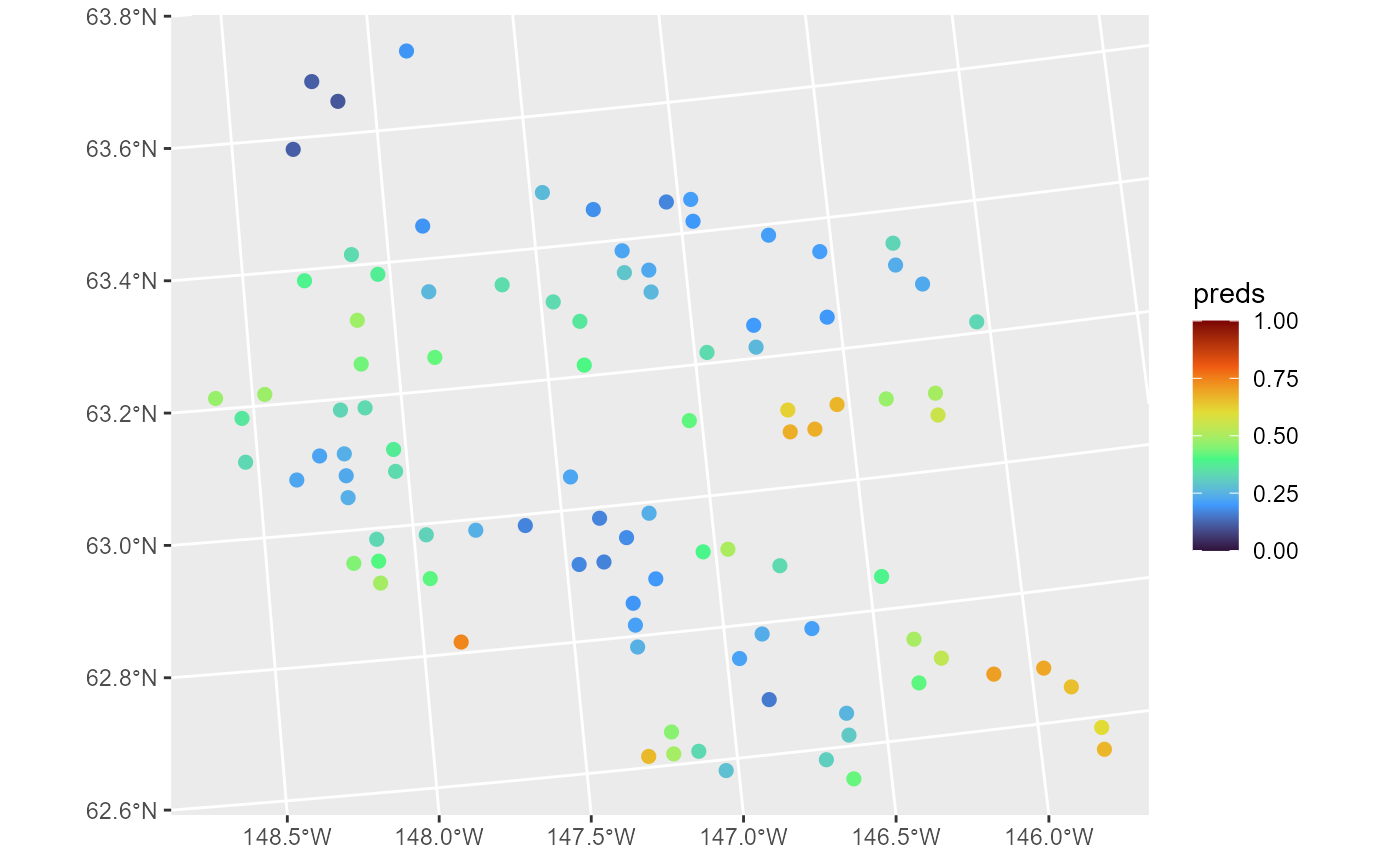
These predictions have similar spatial patterns as moose presence the
observed data. Next we remove the model predictions from
moose_preds and show how augment() can be used
to obtain the same predictions alongside prediction intervals (on the
response scale):
moose_preds$preds <- NULL
augment(binmod, newdata = moose_preds, type.predict = "response", interval = "prediction")#> Simple feature collection with 100 features and 5 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: 269386.2 ymin: 1418453 xmax: 419976.2 ymax: 1541763
#> Projected CRS: NAD83 / Alaska Albers
#> # A tibble: 100 × 6
#> elev strat .fitted .lower .upper geometry
#> * <dbl> <chr> <dbl> <dbl> <dbl> <POINT [m]>
#> 1 143. L 0.705 0.248 0.946 (401239.6 1436192)
#> 2 324. L 0.336 0.0373 0.868 (352640.6 1490695)
#> 3 158. L 0.263 0.0321 0.792 (360954.9 1491590)
#> 4 221. M 0.243 0.0360 0.734 (291839.8 1466091)
#> 5 209. M 0.742 0.270 0.957 (310991.9 1441630)
#> 6 218. L 0.191 0.0196 0.736 (304473.8 1512103)
#> 7 127. L 0.179 0.0226 0.673 (339011.1 1459318)
#> 8 122. L 0.241 0.0344 0.738 (342827.3 1463452)
#> 9 191 L 0.386 0.0414 0.902 (284453.8 1502837)
#> 10 105. L 0.494 0.114 0.882 (391343.9 1483791)
#> # ℹ 90 more rowsFunction Glossary
Here we list some commonly used spmodel functions.
-
AIC(): Compute the AIC. -
AICc(): Compute the AICc. -
anova(): Perform an analysis of variance. -
augment(): Augment data with diagnostics or new data with predictions. -
AUROC(): Compute the area under the receiver operating characteristic curve for binary spatial generalized linear models. -
BIC(): Compute the BIC. -
coef(): Return coefficients. -
confint(): Compute confidence intervals. -
cooks.distance(): Compute Cook’s distance. -
covmatrix(): Return covariance matrices. -
deviance(): Compute the deviance. -
esv(): Compute an empirical semivariogram. -
fitted(): Compute fitted values. -
glance(): Glance at a fitted model. -
glances(): Glance at multiple fitted models. -
hatvalues(): Compute leverage (hat) values. -
logLik(): Compute the log-likelihood. -
loocv(): Perform leave-one-out cross validation. -
model.matrix(): Return the model matrix (\(\mathbf{X}\)). -
plot(): Create fitted model plots. -
predict(): Compute predictions and prediction intervals. -
pseudoR2(): Compute the pseudo r-squared. -
residuals(): Compute residuals. -
spautor(): Fit a spatial linear model for areal data (i.e., spatial autoregressive model). -
spautorRF(): Fit a random forest spatial residual model for areal data. -
spgautor(): Fit a spatial generalized linear model for areal data (i.e., spatial generalized autoregressive model). -
splm(): Fit a spatial linear model for point-referenced data (i.e., geostatistical model). -
splmRF(): Fit a random forest spatial residual model for point-referenced data. -
spglm(): Fit a spatial generalized linear model for point-referenced data (i.e., generalized geostatistical model). -
sprbeta(): Simulate spatially correlated beta random variables. -
sprbinom(): Simulate spatially correlated binomial (Bernoulli) random variables. -
sprgamma(): Simulate spatially correlated gamma random variables. -
sprinvgauss(): Simulate spatially correlated inverse Gaussian random variables. -
sprnbinom(): Simulate spatially correlated negative binomial random variables. -
sprnorm(): Simulate spatially correlated normal (Gaussian) random variables. -
sprpois(): Simulate spatially correlated Poisson random variables. -
summary(): Summarize fitted models. -
tidy(): Tidy fitted models. -
varcomp(): Compare variance components. -
vcov(): Compute variance-covariance matrices of estimated parameters.
For a full list of spmodel functions alongside their
documentation, see the documentation manual. Documentation for methods
of generic functions that are defined outside of spmodel
can be found by running help("generic.spmodel", "spmodel")
(e.g., help("summary.spmodel", "spmodel"),
help("predict.spmodel", "spmodel"), etc.). Note that
?generic.spmodel is shorthand for
help("generic.spmodel", "spmodel").