Note: This article is a work in progress
EXAMPLES OF FILES & TEST DATA EJAM CAN IMPORT OR OUTPUT
Sample spreadsheets & shapefiles for trying the web app
Examples of .xlsx files and shapefiles are installed locally with EJAM, as input files you can use to try out EJAM functions or the web app, or to see what an input file should look like.
Local folders with sample files
The best, simplest way to see all these files is the function called testdata()
testdata()For more details, you might find the following useful, but testdata() will probably provide what you are looking for.
Another way to see a list of local folders (where EJAM is installed locally) with files that can be uploaded to the EJAM web app or provided as inputs to EJAM functions (as examples or for testing):
x <- list.dirs(system.file("testdata/", package = "EJAM"), recursive = FALSE)
cbind(`Local folders with installed file examples` = x, Subfolder = basename(x))To see a list of the files available locally as installed sample/test data:
for (example_type in x) {
cat("\n", basename(example_type), "\n\n")
these = matrix(list.files(example_type))
colnames(these) <- paste0("Examples Installed in /testdata/", basename(example_type), "/")
print(cbind(these))
}
#>
#> address
#>
#> Examples Installed in /testdata/address/
#> [1,] "street_address_9.xlsx"
#> [2,] "test_address_table.xlsx"
#> [3,] "test_address_table_9.xlsx"
#> [4,] "test_address_table_goodnames.xlsx"
#> [5,] "test_address_table_withfull.xlsx"
#>
#> ejscreenapi
#>
#> Examples Installed in /testdata/ejscreenapi/
#> [1,] "datafile_apiv2.3_output_example.pjson"
#> [2,] "datafile_apiv2.3_output_example_json.R"
#>
#> examples_of_output
#>
#> Examples Installed in /testdata/examples_of_output/
#> [1,] "testoutput_doaggregate_1000pts_1miles.xlsx"
#> [2,] "testoutput_doaggregate_100pts_1miles.xlsx"
#> [3,] "testoutput_doaggregate_10pts_1miles.xlsx"
#> [4,] "testoutput_ejamit2excel_1000pts_1miles.xlsx"
#> [5,] "testoutput_ejamit2excel_100pts_1miles.xlsx"
#> [6,] "testoutput_ejamit2excel_10pts_1miles.xlsx"
#> [7,] "testoutput_report_10pts_1Miles.html"
#>
#> fips
#>
#> Examples Installed in /testdata/fips/
#> [1,] "cities_2.xlsx"
#> [2,] "counties_in_AL_detailed.xlsx"
#> [3,] "counties_in_Alabama.xlsx"
#> [4,] "counties_in_Delaware.xlsx"
#> [5,] "counties_in_Delaware_invalid.xlsx"
#> [6,] "county_10.xlsx"
#> [7,] "county_100.xlsx"
#> [8,] "county_1000.xlsx"
#> [9,] "county_state_300.xlsx"
#> [10,] "state_10.xlsx"
#> [11,] "state_50.xlsx"
#> [12,] "state_county_tract_10.xlsx"
#> [13,] "tract_10.csv"
#> [14,] "tract_100.csv"
#> [15,] "tract_1000.csv"
#> [16,] "tract_state_285.xlsx"
#>
#> latlon
#>
#> Examples Installed in /testdata/latlon/
#> [1,] "3points.xlsx"
#> [2,] "testpoints_02.xlsx"
#> [3,] "testpoints_10.xlsx"
#> [4,] "testpoints_100.xlsx"
#> [5,] "testpoints_100_sites_with_signif_violations_NAICS_326_ECHO.xlsx"
#> [6,] "testpoints_1000.xlsx"
#> [7,] "testpoints_1000_Latitude_LONG.xlsx"
#> [8,] "testpoints_10000.xlsx"
#> [9,] "testpoints_100000.xlsx"
#> [10,] "testpoints_2.xlsx"
#> [11,] "testpoints_207_sites_with_signif_violations_NAICS_326_ECHO.csv"
#> [12,] "testpoints_5.xlsx"
#> [13,] "testpoints_50.xlsx"
#> [14,] "testpoints_500.xlsx"
#> [15,] "testpoints_bad.xlsx"
#> [16,] "testpoints_invalid_latlon.xlsx"
#> [17,] "testpoints_invalid_latlon_more_cases.xlsx"
#> [18,] "testpoints_overlap3.xlsx"
#> [19,] "testpoints_PR_GU_AS_VI_MP.xlsx"
#>
#> program_type
#>
#> Examples Installed in /testdata/program_type/
#> [1,] "program_name_only_3.xlsx"
#>
#> programid
#>
#> Examples Installed in /testdata/programid/
#> [1,] "program_test_data_10.csv"
#> [2,] "program_test_data_10.xlsx"
#> [3,] "program_test_data_100.csv"
#> [4,] "program_test_data_1000.csv"
#> [5,] "program_test_data_10000.csv"
#> [6,] "test_pgm_sys_id_1000.xlsx"
#>
#> registryid
#>
#> Examples Installed in /testdata/registryid/
#> [1,] "ECHO_Test_Data"
#> [2,] "FRS_example_data.CSV"
#> [3,] "FRS_example_data.xlsx"
#> [4,] "frs_test_regid_8.xlsx"
#> [5,] "frs_testpoints_10.csv"
#> [6,] "frs_testpoints_10.xlsx"
#> [7,] "frs_testpoints_100.csv"
#> [8,] "frs_testpoints_100.xlsx"
#> [9,] "frs_testpoints_1000.csv"
#> [10,] "frs_testpoints_1000.xlsx"
#> [11,] "frs_testpoints_10000.csv"
#> [12,] "frs_testpoints_10000.xlsx"
#> [13,] "frs_testpoints_100000.csv"
#> [14,] "frs_testpoints_100000.xlsx"
#> [15,] "frs_testpoints_3_duplicated_id.xlsx"
#> [16,] "testids_registry_id_8.xlsx"
#>
#> shapes
#>
#> Examples Installed in /testdata/shapes/
#> [1,] "portland.gdb"
#> [2,] "portland.gdb.zip"
#> [3,] "portland.json"
#> [4,] "portland_folder_shp"
#> [5,] "portland_folder_shp.zip"
#> [6,] "portland_shp.zip"
#> [7,] "stations.zip"
#> [8,] "stations_shp.zip"
#> [9,] "testshapes_2.zip"You can try uploading these kinds of files in the web app, for example, by finding them in these local folders where you installed the package:
- /
EJAM/testdata/latlon/testpoints_100.xlsx - /
EJAM/testdata/shapes/portland_shp.zip - etc.
To open the locally installed “testdata” folders (in Windows File Explorer, or MacOS Finder)
browseURL(system.file("testdata", package = "EJAM"))Example of using a file in EJAM
testpoint_files <- list.files(
system.file("testdata/latlon", package = "EJAM"),
full.names = T
)
testpoint_files
latlon_from_anything(testpoint_files[2]) Sample R data objects: Examples of inputs & outputs of EJAM functions
The package has a number of data objects, installed as part of EJAM and related packages, that are examples of inputs or intermediate data objects that you can use to try out EJAM functions, or you may just want to see what the outputs and inputs look like, or you could use them for testing purposes.
For documentation on each input or output item (R object), see https://usepa.github.io/EJAM/reference/index.html#test-data
This code snippet provides a useful list of test/ sample data objects in EJAM and related packages:
POINT DATA (LAT/LON COORDINATES) for testing ejamit(), mapfast(), ejscreenit(), getblocksnearby(), etc.
x[grepl("^testp", x$Item), ]
#> Package Item
#> 102 EJAM testpoints_10
#> 124 EJAM testpoints_100
#> 122 EJAM testpoints_100_dt
#> 127 EJAM testpoints_1000
#> 156 EJAM testpoints_10000
#> 110 EJAM testpoints_5
#> 120 EJAM testpoints_50
#> 137 EJAM testpoints_500
#> 111 EJAM testpoints_bad
#> 103 EJAM testpoints_overlap3
#> Title
#> 102 test points data.frame with columns sitenumber, lat, lon
#> 124 test points data.frame with columns sitenumber, lat, lon
#> 122 test points data.frame with columns sitenumber, lat, lon
#> 127 test points data.frame with columns sitenumber, lat, lon
#> 156 test points data.frame with columns sitenumber, lat, lon
#> 110 test points data.frame with columns sitenumber, lat, lon
#> 120 test points data.frame with columns sitenumber, lat, lon
#> 137 test points data.frame with columns sitenumber, lat, lon
#> 111 test points data.frame with columns note, lat, lon
#> 103 test points data.frame with columns note, lat, lonSTREET ADDRESSES for testing geocoding in latlon_from_address() etc.
x[grepl("^test_", x$Item), ]
#> Package Item
#> 100 EJAM test_address_parts1
#> 107 EJAM test_address_table
#> 121 EJAM test_address_table_9
#> 108 EJAM test_address_table_goodnames
#> 109 EJAM test_address_table_withfull
#> 101 EJAM test_addresses_9
#> 43 EJAM test_addresses2
#> 3 EJAM test_regid
#> 4 EJAM test_xtrac
#> Title
#> 100 datasets for trying address-related functions
#> 107 datasets for trying address-related functions
#> 121 datasets for trying address-related functions
#> 108 datasets for trying address-related functions
#> 109 datasets for trying address-related functions
#> 101 datasets for trying address-related functions
#> 43 datasets for trying address-related functions
#> 3 test_regid (DATA) test data, vector of EPA FRS Registry ID numbers
#> 4 for internal use
cat("\n\n")FACILITY REGISTRY IDs for testing latlon_from_regid() etc.
x[grepl("^test[^op_]", x$Item), ]
#> Package Item
#> 9 EJAM testids_program_sys_id
#> 5 EJAM testids_registry_id
#> 126 EJAM testshapes_2
#> Title
#> 9 test data, string vector of EPA FRS Program System ID numbers
#> 5 test data, vector of EPA FRS Registry ID numbers
#> 126 testshapes_2 dataset
cat("\n\n")EXAMPLES OF OUTPUTS from ejamit(), ejscreenit(), getblocksnearby(), etc., you can use as inputs to ejam2report(), ejam2excel(), ejam2ratios(), ejam2barplot(), doaggregate(), etc.
x[grepl("^testout", x$Item), ]
#> Package Item
#> 165 EJAM testoutput_doaggregate_1000pts_1miles
#> 157 EJAM testoutput_doaggregate_100pts_1miles
#> 148 EJAM testoutput_doaggregate_10pts_1miles
#> 166 EJAM testoutput_ejamit_1000pts_1miles
#> 158 EJAM testoutput_ejamit_100pts_1miles
#> 151 EJAM testoutput_ejamit_10pts_1miles
#> 134 EJAM testoutput_ejscreenapi_1pts_1miles
#> 140 EJAM testoutput_ejscreenapi_plus_5
#> 146 EJAM testoutput_ejscreenit_10pts_1miles
#> 144 EJAM testoutput_ejscreenit_5
#> 152 EJAM testoutput_ejscreenit_50
#> 162 EJAM testoutput_ejscreenit_500
#> 129 EJAM testoutput_ejscreenRESTbroker_1pts_1miles
#> 161 EJAM testoutput_getblocksnearby_1000pts_1miles
#> 149 EJAM testoutput_getblocksnearby_100pts_1miles
#> 139 EJAM testoutput_getblocksnearby_10pts_1miles
#> Title
#> 165 test output of doaggregate()
#> 157 test output of doaggregate()
#> 148 test output of doaggregate()
#> 166 test output of ejamit()
#> 158 test output of ejamit()
#> 151 test output of ejamit()
#> 134 test data, output from this function
#> 140 test data examples of output from 'ejscreenapi_plus()' using testpoints_5, radius = 1
#> 146 test output of ejscreenit(), using the EJScreen API
#> 144 test data examples of output from 'ejscreenit()' using testpoints_5, radius = 1
#> 152 test data examples of output from 'ejscreenit()' using testpoints_50, radius = 1
#> 162 test data examples of output from 'ejscreenit()' using testpoints_500, radius = 1
#> 129 test data, output from this function
#> 161 test output of getblocksnearby(), and is an input to doaggregate()
#> 149 test output of getblocksnearby(), and is an input to doaggregate()
#> 139 test output of getblocksnearby(), and is an input to doaggregate()
cat("\n\n")LARGE DATASETS USED BY THE PACKAGE
Note that the largest files used by the package are mostly the block-related datasets with info about population size and location of US blocks, the facility datasets with info about EPA-regulated sites, and the blockgroup-related datasets with EJScreen indicators.
Some datasets get downloaded by the package at installation or launch or as needed. Datasets may include for example: [blockwts], [blockpoints], [quaddata], [blockid2fips], [frs], [frs_by_programid], [frs_by_naics], [frs_by_sic], and [frs_by_mact].
Blockgroup-related datasets incude [blockgroupstats], [bgpts], [bgej], [usastats], [statestats], [bgid2fips], and [bg_cenpop2020]. For more info see https://usepa.github.io/EJAM/reference/index.html#datasets-with-indicators-etc-
USING NAICS AND SIC CODES TO LOCATE FACILITIES BY INDUSTRY
EJAM helps select regulated sites based on industrial classification, using NAICS or SIC code. Finding the right NAICS and finding all the right sites by NAICS is complicated. Doing so requires understanding the NAICS system and the FRS dataset, and the functions in EJAM that help find or use NAICS codes.
NAICS/SIC categories can be explored in a few ways: - NAICS.com website with extensive information about NAICS and SIC - Key EJAM functions for using NAICS/SIC - EPA FRS Facility Industrial Classification Search tool where you can find facilities based on NAICS or SIC. - EPA APIs exist that can be used for similar queries.
Some key EJAM functions include [regid_from_naics()], [latlon_from_naics()], [frs_from_naics()], [naics_findwebscrape()], and [naics_categories()]. These functions can help find EPA FRS sites based on naics codes or titles. They rely on [frs_by_naics] (a data.table), and [naics_from_any()] for querying by code or title of category.
Important points:
Note that a very large fraction of all FRS sites (as obtained for use in EJAM) lack NAICS code!
Note that EJAM may query FRS sites differently than the FRS search tool or other query tools would.
Note that NAICS.com reports many more businesses for a given 6-digit category than the FRS shows, which might be due to FRS only including EPA-regulated sites but also due to data gaps.
Note the difference between children = TRUE and children = FALSE in EJAM functions like latlon_from_naics()
Note that searching on a 6-digit code misses parent categories you may want. The FRS data on NAICS by site is inconsistent in how many digits are reported for the NAICS (explained below).
A given site might be listed in the FRS as being under one or more NAICS codes of various lengths, such as only a parent code (large grouping), only a detailed code (6-digit), or some combination of codes and their subcategories.
And the same title, like “Petroleum Refineries,” may be assigned by the NAICS system to the category but also a subcategory, as with codes 32411 and 324110. The function [naics_from_any()] shows what codes and title exist in the NAICS system.
Also, certain terms appear in the online description of a NAICS but not in the title of the NAICS – the function [naics_findwebscrape()] helps with those cases, e.g., compare these:
naics_findwebscrape("cement")
naics_from_any("cement")Compare also these:
naics_findwebscrape("refiner")
naics_from_any("refiner")naics_findwebscrape("refiner") reports “324110”
(Petroleum Refineries) and other related industries, but not the 5-digit
“32411” (also Petroleum Refineries).
naics_from_any("refiner") reports “324110” and “32411”
but not other related industries.
Using naics_findwebscrape() finds only the 6-digit codes
that match on title or description, so it would find some codes not
found by [naics_from_any()] which does not query description, but could
lead to missing some facilities in the sense that the 6-digit code does
not cover the sites listed in FRS under only the 5-digit code for
Petroleum Refineries (not the 6-digit).
It is important to note that searching on a 6-digit code misses parent categories that may include sites you expect to find:
frs_from_naics("324110", children = F)[,1:5] finds a few
hundred sites, but it fails to find some sites you would find using
frs_from_naics("32411", children = F)[,1:5]
The code example below shows that the FRS dataset has some facilities listed under the 5-digit “32411” code only, some with the 6-digit “324110” code only, and some with both codes:
hasboth = intersect(
frs_from_naics("32411", children = F)[,1:5]$REGISTRY_ID,
frs_from_naics("324110", children = F)[,1:5]$REGISTRY_ID
)
hasonly6digit = setdiff(
frs_from_naics("32411", children = F)[,1:5]$REGISTRY_ID,
frs_from_naics("324110", children = F)[,1:5]$REGISTRY_ID
)
hasonly5digit = setdiff(
frs_from_naics("324110", children = F)[,1:5]$REGISTRY_ID,
frs_from_naics("32411", children = F)[,1:5]$REGISTRY_ID
)
length(hasonly5digit) # Most of the FRS sites here
#> [1] 362
length(hasonly6digit)
#> [1] 12
length(hasboth)
#> [1] 12Examples of some NAICS/SIC functions
naics_from_any(naics_categories(3))[order(name),.(name,code)][1:10,]
naics_from_any(naics_categories(3))[order(code),.(code,name)][1:10,]
naics_from_code(211)
naicstable[code==211,]
naics_subcodes_from_code(211)
naics_from_code(211, children = TRUE)
naicstable[n3==211,]
NAICS[211][1:3] # wrong
NAICS[NAICS == 211]
NAICS["211 - Oil and Gas Extraction"]
naics_from_any("plastics and rubber")[,.(name,code)]
naics_from_any(326)
naics_from_any(326, children = T)[,.(code,name)]
naics_from_any("plastics", children=T)[,unique(n3)]
naics_from_any("pig")
naics_from_any("pig ") # space after g
# naics_from_any("copper smelting")
# naics_from_any("copper smelting", website_scrape=TRUE)
# browseURL(naics_from_any("copper smelting", website_url=TRUE) )
a = naics_from_any("plastics")
b = naics_from_any("rubber")
fintersect(a,b)[,.(name,code)] # a AND b
funion(a,b)[,.(name,code)] # a OR b
naics_subcodes_from_code(funion(a,b)[,code])[,.(name,code)] # plus children
naics_from_any(funion(a,b)[,code], children=T)[,.(name,code)] # same
NROW(naics_from_any(325))
#[1] 1
NROW(naics_from_any(325, children = T))
#[1] 54
NROW(naics_from_any("chem"))
#[1] 20
NROW(naics_from_any("chem", children = T))
# [1] 104HOW TO ANALYZE PROXIMITY USING EJAM
An outline of how to use key functions is provided below. After these examples is a discussion of background information and considerations in selecting radius.
RESIDENTIAL POPULATION GROUP PERCENTAGES BY DISTANCE AT BLOCK GROUP RESOLUTION
It is easiest to analyze distance increments based on each blockgroup’s average resident here. Block resolution is covered in a later section.
WITHIN ONE RADIUS
Overall list of sites
At the OVERALL LIST of sites as a whole, which groups are overrepresented within X mile radius vs Statewide?
out <- ejamit(testpoints_100, radius = 3.1)
ejam2ratios(out)
#>
#>
#> Average Resident in Place(s) Analyzed vs US or State
#> Ratio_to_US_avg Ratio_to_State_avg
#> Demog.Ind. 1.2 1.2
#> Suppl Demog.Ind. 1.0 1.1
#> %Low-inc. 1.0 1.1
#> %Limited English 1.8 1.4
#> %Unemployed 1.2 1.1
#> %< High School 1.2 1.2
#> %< age 5 1.1 1.1
#> %> age 64 0.9 0.9
#> %POC 1.5 1.3
#> %Hispanic 1.6 1.3
#> %Black NHA 1.2 1.5
#> %Asian NHA 2.1 1.3
#> %AmerIndian/AK NHA 0.4 0.8
#> %Hawaiian/PI NHA 0.9 1.2
#> %Other race NHA 1.6 1.2
#> %multirace NH 1.0 1.0
#> %White NHA 0.7 0.8
#> PM2.5 1.1 1.0
#> Ozone 1.1 1.0
#> NO2 1.6 1.3
#> Diesel PM 2.2 1.6
#> Toxic Releases to Air 0.5 1.2
#> Traffic 2.5 1.6
#> %pre-1960 1.7 1.2
#> NPL 2.8 1.5
#> RMP 1.2 1.3
#> TSDF 3.6 1.9
#> UST 3.2 2.0
#> NPDES 0.0 0.7
#> Drinking 1.8 1.6
ejam2barplot(out)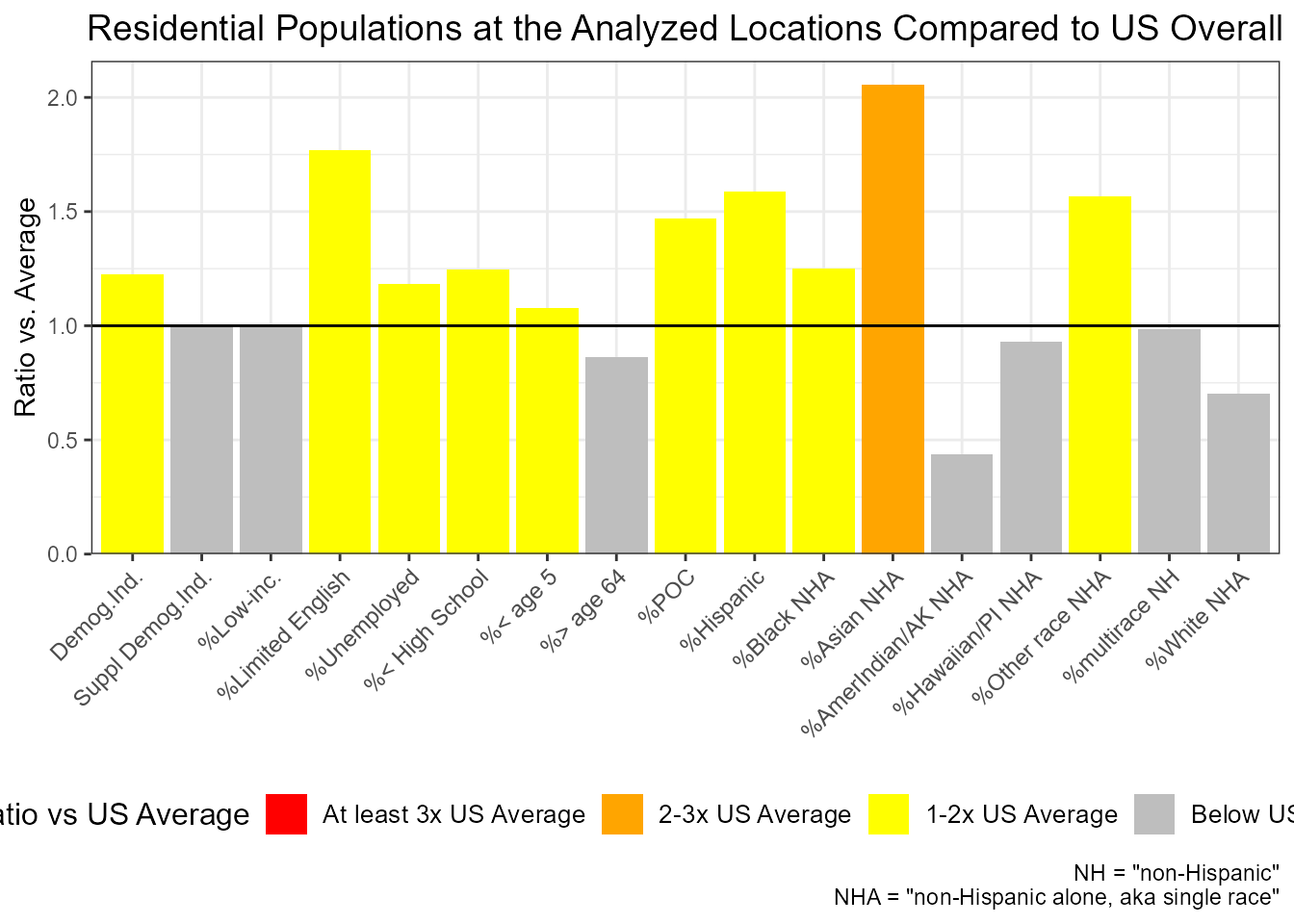
Example of ejam2barplot() showing percent Asian among residents within 5 km of these 100 sites is more than two times the US rate overall
Just one site
At JUST ONE SITE, which groups are overrepresented within X mile radius vs Statewide?
out1 <- ejamit(testpoints_100[2, ], radius = 3.1)
ejam2ratios(out1)
ejam2barplot(out1)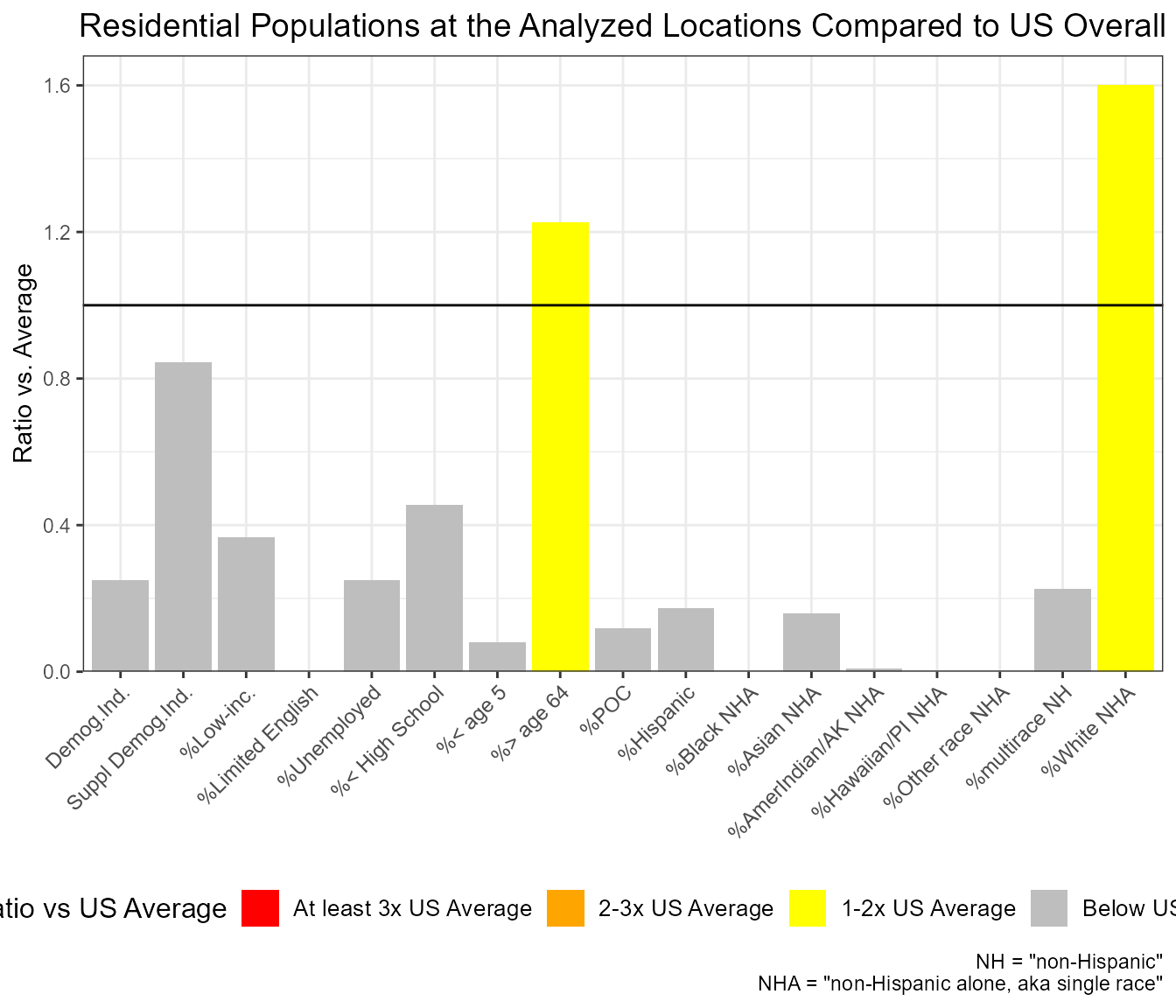
Example of ejam2barplot() showing percent non-Hispanic White Alone among residents within 5 km of this one site is about 1.6 times the US rate overall
Site by site comparison
Which groups are overrepresented at EACH SITE, within X mile radius vs Statewide
out <- testoutput_ejamit_10pts_1miles
x = round(data.frame(out$results_bysite)[, c("ratio.to.state.avg.pctlowinc", "ratio.to.state.avg.pctmin")], 2)
names(x) = fixcolnames(names(x),"r","shortlabel")
x = data.frame(sitenumber = 1:NROW(x), x)
x
#> sitenumber Ratio.to.State.avg..Low.inc. Ratio.to.State.avg..POC
#> 1 1 0.97 0.54
#> 2 2 1.08 0.99
#> 3 3 0.57 0.44
#> 4 4 0.33 0.72
#> 5 5 2.12 1.59
#> 6 6 2.22 1.76
#> 7 7 0.97 1.35
#> 8 8 0.68 0.87
#> 9 9 1.08 1.12
#> 10 10 0.27 0.44Plot to compare sites, for just one residential population indicator
This plot shows that % low income among residents at sites 5 and 6 is more than twice the relevant State average. It is near average at several other sites, and is less than half the State average at sites 4 and 10.
ejam2barplot_sites(out, "ratio.to.state.avg.pctlowinc", topn = 10, sortby = F)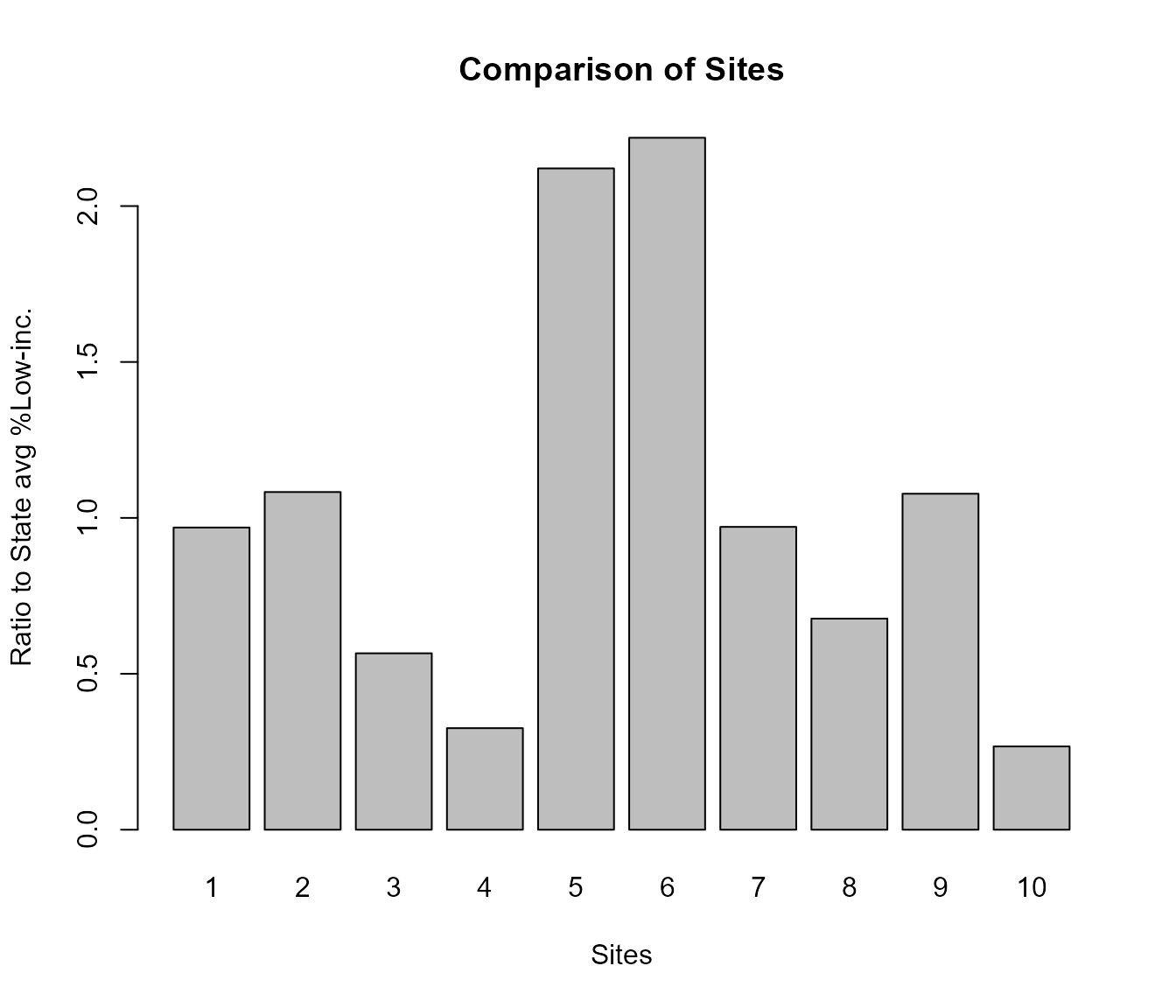
Example of ejam2barplot_sites()
## For raw values at key sites:
# ejam2barplot_sites(out, "pctlowinc")WITHIN MULTIPLE DISTANCES - COMPARING RADIUS CHOICES
Overall list of sites
At the OVERALL LIST of sites as a whole, which groups are overrepresented within X mile radius vs Statewide?
radii <- c(1,2,3,10)
#radii <- c(1, 10) # quicker example
pts <- testpoints_100[10:12, ]See just the table
x <- ejamit_compare_distances(pts, radii = radii, quiet = TRUE, plot = FALSE)#> Analyzing 3 points, radius of 1 miles around each.
#> Analyzing 3 points, radius of 2 miles around each.
#> Analyzing 3 points, radius of 3 miles around each.
#> Analyzing 3 points, radius of 10 miles around each.
#>
#> 1 2 3 10
#> Ratio to State avg %Hispanic 2.5 2.3 2.3 1.8
#> Ratio to State avg %Black NHA 0.4 0.2 0.2 1.2
#> Ratio to State avg %Asian NHA 0.1 0.4 0.5 1.0
#> Ratio to State avg %AmerIndian/AK NHA 0.3 0.2 0.2 0.5
#> Ratio to State avg %Hawaiian/PI NHA 0.0 0.0 0.1 0.6
#> Ratio to State avg %Other race NHA 0.7 1.1 0.7 0.8
#> Ratio to State avg %multirace NH 0.1 0.1 0.1 0.4
#> Ratio to State avg %White NHA 0.2 0.3 0.2 0.3See the plot
# x <- ejamit_compare_distances(pts, radii = radii, quiet = TRUE) # in which default is plot=TRUE
# or
ejamit_compare_distances2plot(x)
#>
#> Indicators that increase the most as you get closer: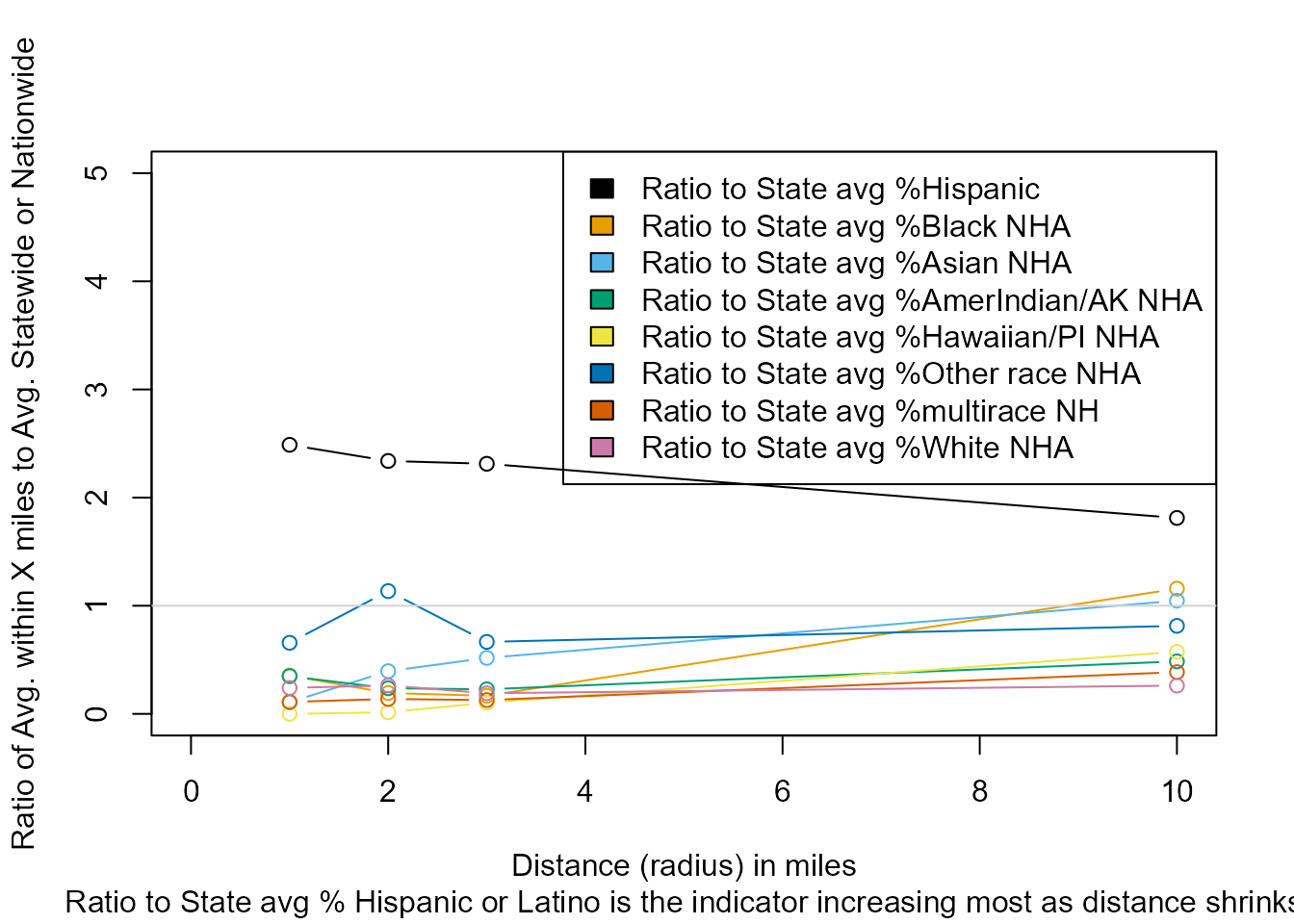
Example of using ejamit_compare_distances2plot()
#> [1] "Ratio to State avg % Hispanic or Latino"RESIDENTIAL POPULATION GROUP DATA AT BLOCK GROUP RESOLUTION
Most of the EJAM functions use distance to the average resident of a block group, which is calculated from the distance to each block’s internal point and uses the approximation that within a block the average resident and all residents are as far as that internal point. For typical distances analyzed in EJAM (e.g., 3 mile radius, or about 5 km) that is a good approximation, since only about 2% of all US blocks are larger than 1 square mile.
If you need high spatial resolution (block by block) plots of an indicator as a function of distance, you can directly work with getblocksnearby() or just use the function plot_distance_by_pctd(). It uses the distance from the site to each block’s internal point (like a centroid) rather than just the distance to the average resident in each block group.
How residential population group percentages at ONE SITE vary as a continuous function of distance
Example of area where %Black is very high within 1 mile but drops by 3 miles away
pts <- testpoints_100[3,]
y <- plot_distance_by_pctd(
getblocksnearby(pts, radius = 10, quiet = T),
score_colname = "pctnhba",
sitenumber = 1)
#> Analyzing 1 points, radius of 10 miles around each.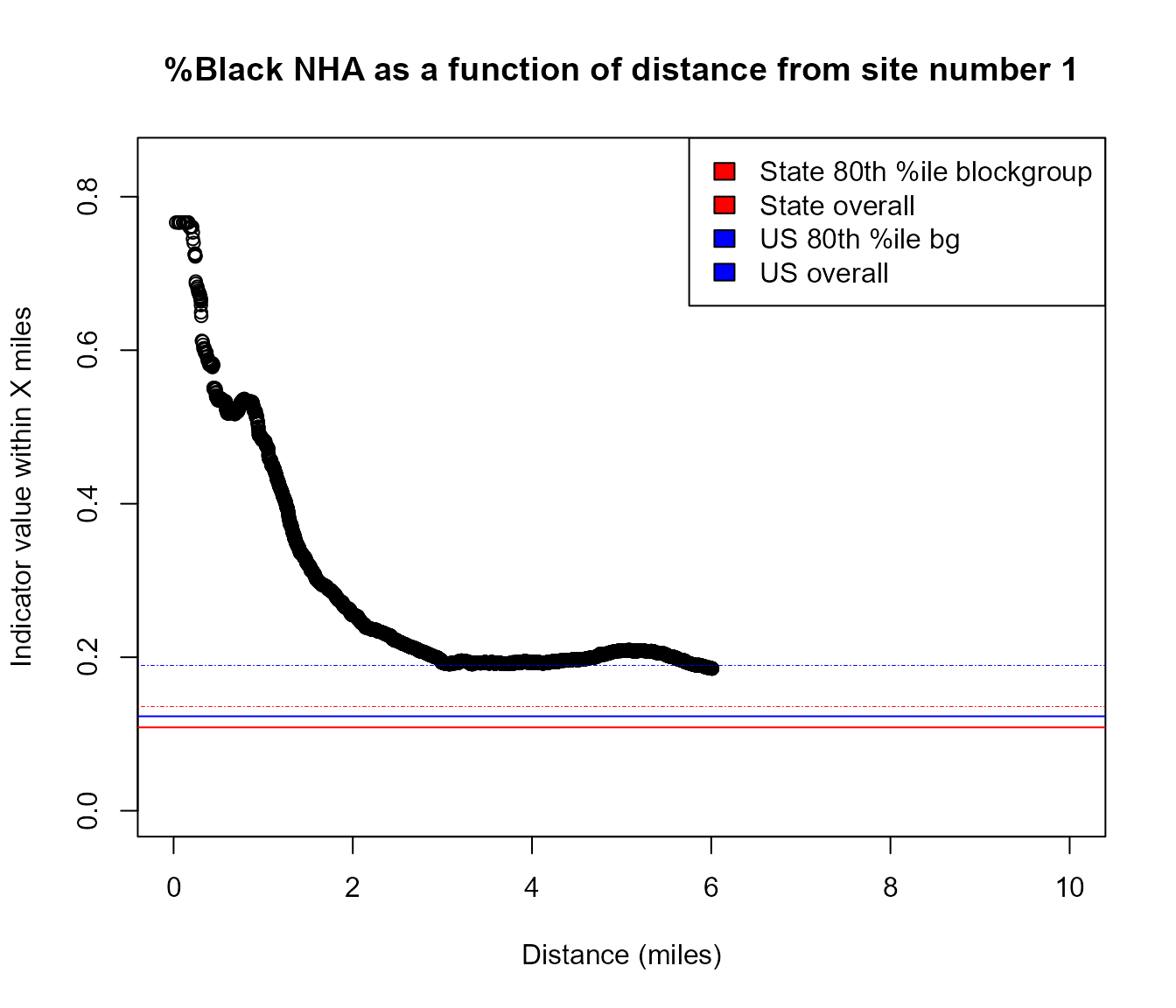
Example of using plot_distance_by_pctd()
#browseURL(url_ejscreen_report(lat = pts$lat, lon = pts$lon, radius = 0.5))
#browseURL(url_ejscreen_report(lat = pts$lat, lon = pts$lon, radius = 3))Example of area that has higher %Hispanic as you go 10 to 30 miles away from this specific point
pts <- data.table::data.table(lat = 45.75464, lon = -94.36791)
y <- plot_distance_by_pctd(pts,
sitenumber = 1, score_colname = "pcthisp")
#> Analyzing 1 points, radius of 30 miles around each.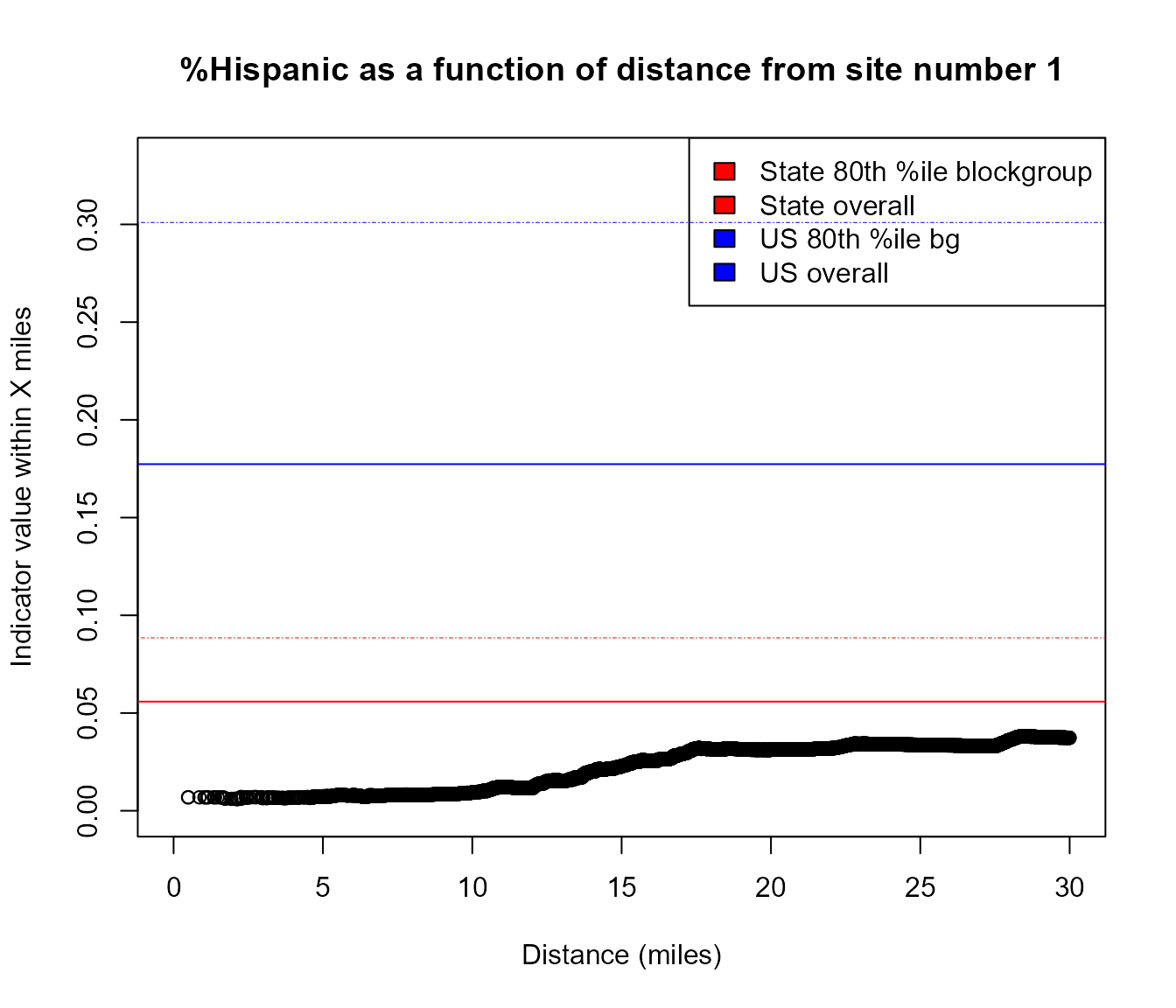
Example of using plot_distance_by_pctd()
# browseURL(url_ejscreen_report(lat = pts$lat, lon = pts$lon, radius = 10))
# browseURL(url_ejscreen_report(lat = pts$lat, lon = pts$lon, radius = 30))Step through all the sites to see an indicator versus distance at each
Examples of sites analyzed here show some conclusions are very sensitive to the radius used. The choice of radius in proximity analysis for some sites will lead to a very different conclusion depending on the radius analyzed, if only a single distance is checked or reported on. The relationship between distance X and percent by residential population group within X miles can be positive, negative, or roughly flat, etc., depending on the site and group. The residential population group percentage may be above or below the US average or the State average within a given distance of the site.
For the ten sites analyzed in this example, a wide range of patterns is found:
At site 5, % low income is extremely high very close to the site and falls sharply with distance but it remains quite high (still above 80th percentile of US or State) even within 4 miles.
At site number 2 here, % low income very close to the site is around the 80th percentile in the State, and is around the US 80th percentile within about 1 mile, but then it falls to below State and then US average within around 2 and then 3 miles of the site.
At site 7, it is below average until about 8 miles, but is above US and State averages within 10 miles.
At site 9, it can be above or below average in State and/or in US, depending on the distance, but it is never as high as the 80th percentiles.
At sites 2, 3, 4, and 10, % low income is far below US and State averages within any distance shown here.
pts <- testpoints_10
s2b <- getblocksnearby(pts, radius = 10, quiet = T)
for (i in 1:NROW(pts)) {
plot_distance_by_pctd(s2b, sitenumber = i, score_colname = "pctlowinc")
readline() # hit any key to step through the plots
}Block by block details are also easy to view in a map of all the nearby blocks, as shown in the section on [plotblocksnearby()] and details of blocks near one site.
Cumulative Distribution plots of groups as a continuous function of distance
Out of all the residents within the area analyzed, see how some are mostly nearby and others are further away, as a CDF plot. This shows the share of each residential population group residing at various distances from sites, with distance from nearest site on the x axis and the cumulative share of each group on the y axis (of all residents within 10 miles, what percent have a site within X miles?). It compares everyone nearby to just those who are among the percent low income, and shows that, for example, a larger share of all the low income population within 10 miles actually live within about 6 miles than is the case for everyone within 10 miles. In other words, within the 10 mile radius circles, more of the low income residents are closer to a site than are the non-low income residents or all residents.
# out <- ejamit(testpoints_10, radius = 10)
distance_by_group_plot(
out$results_bybg_people,
demogvarname = 'pctlowinc', demoglabel = 'Low Income')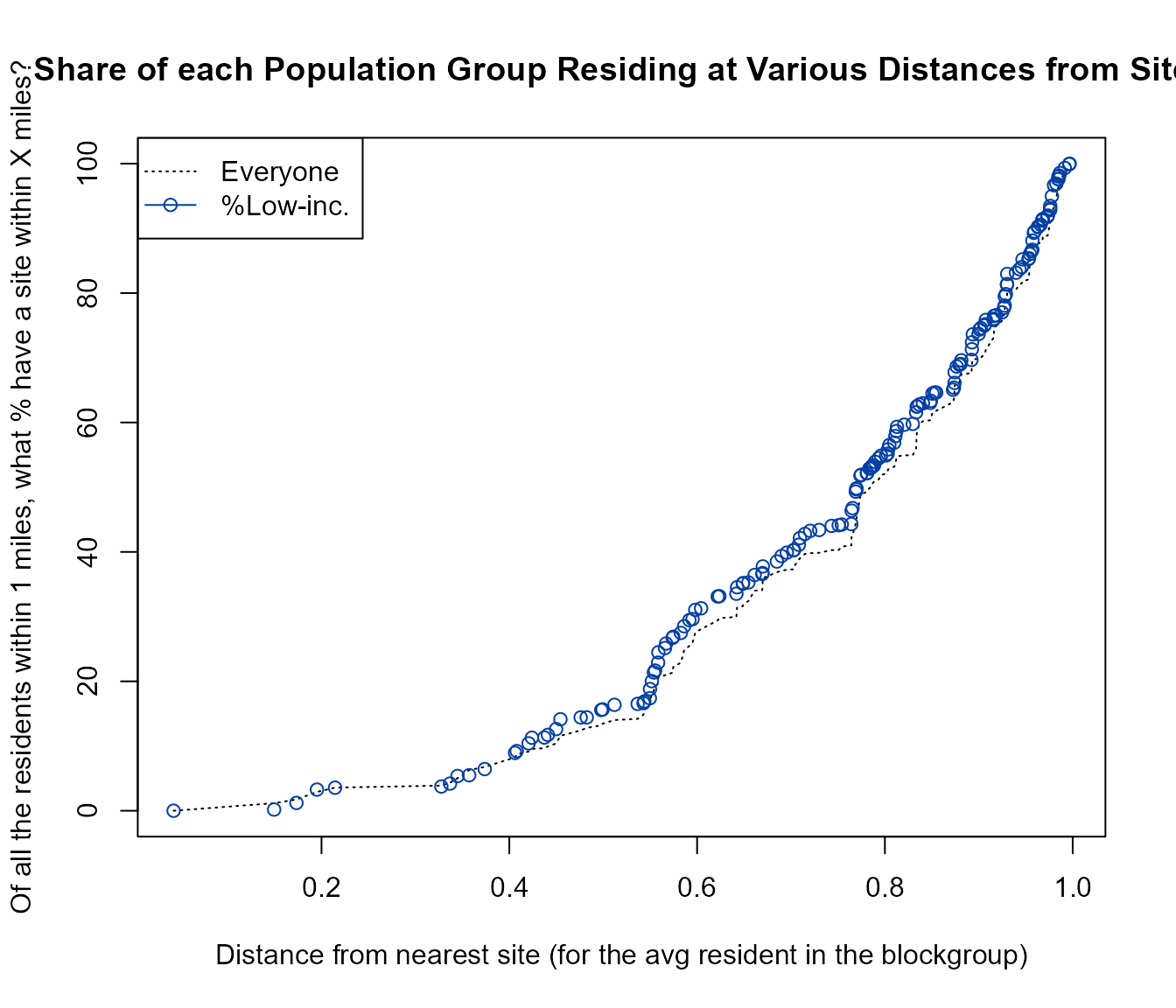
Example of using distance_by_group_plot()
MEAN DISTANCE BY RESIDENTIAL POPULATION GROUP
The analysis described above looks at residential population group percentages as a function of distance. Another perspective is provided by looking at distance as a function of residential population group. This means looking at the average distance or the whole distribution of distances (or proximities) among all the residents within a single residential population group, one group at a time, and comparing these groups.
Overall list of sites
Mean distance of each group, at the OVERALL LIST of sites as a whole
To see a table of residential population indicators, showing the mean distance for each group, compared to distance for those not in that residential population group:
out <- testoutput_ejamit_1000pts_1miles
## But try a larger radius to reveal more information:
# out <- ejamit(testpoints_100, radius = 10)
# see a table of demog indicators
distance_mean_by_group(out$results_bybg_people)
#> group nearest nearer ratio avg_distance_for_group
#> Demog.Ind. Demog.Index FALSE FALSE 1.010 0.69
#> Suppl Demog.Ind. Demog.Index.Supp FALSE FALSE 1.010 0.68
#> %Low-inc. pctlowinc FALSE TRUE 0.984 0.68
#> %Limited English pctlingiso FALSE TRUE 0.987 0.68
#> %Unemployed pctunemployed FALSE TRUE 0.990 0.68
#> %< High School pctlths FALSE TRUE 0.986 0.68
#> %< age 5 pctunder5 FALSE FALSE 1.004 0.69
#> %> age 64 pctover64 FALSE FALSE 1.002 0.69
#> %POC pctmin FALSE TRUE 0.998 0.69
#> %Hispanic pcthisp FALSE TRUE 0.994 0.69
#> %Black NHA pctnhba FALSE FALSE 1.001 0.69
#> %Asian NHA pctnhaa FALSE FALSE 1.007 0.69
#> %AmerIndian/AK NHA pctnhaiana TRUE TRUE 0.930 0.64
#> %Hawaiian/PI NHA pctnhnhpia FALSE TRUE 0.983 0.68
#> %Other race NHA pctnhotheralone FALSE FALSE 1.021 0.70
#> %multirace NH pctnhmulti FALSE FALSE 1.007 0.69
#> %White NHA pctnhwa FALSE FALSE 1.002 0.69
#> avg_distance_for_nongroup
#> Demog.Ind. 0.68
#> Suppl Demog.Ind. 0.68
#> %Low-inc. 0.69
#> %Limited English 0.69
#> %Unemployed 0.69
#> %< High School 0.69
#> %< age 5 0.69
#> %> age 64 0.69
#> %POC 0.69
#> %Hispanic 0.69
#> %Black NHA 0.69
#> %Asian NHA 0.69
#> %AmerIndian/AK NHA 0.69
#> %Hawaiian/PI NHA 0.69
#> %Other race NHA 0.69
#> %multirace NH 0.69
#> %White NHA 0.69
# for just 1 indicator
print(distance_mean_by_group(
out$results_bybg_people,
demogvarname = 'pctlowinc', demoglabel = 'Low Income'))
#> group nearest nearer ratio avg_distance_for_group
#> Low Income pctlowinc TRUE TRUE 0.984 0.68
#> avg_distance_for_nongroup
#> Low Income 0.69To see a barplot, comparing just race/ethnicity groups:
plot_distance_mean_by_group(out$results_bybg_people,
demogvarname = names_d_subgroups,
demoglabel = fixcolnames(names_d_subgroups, "r", "shortlabel")
)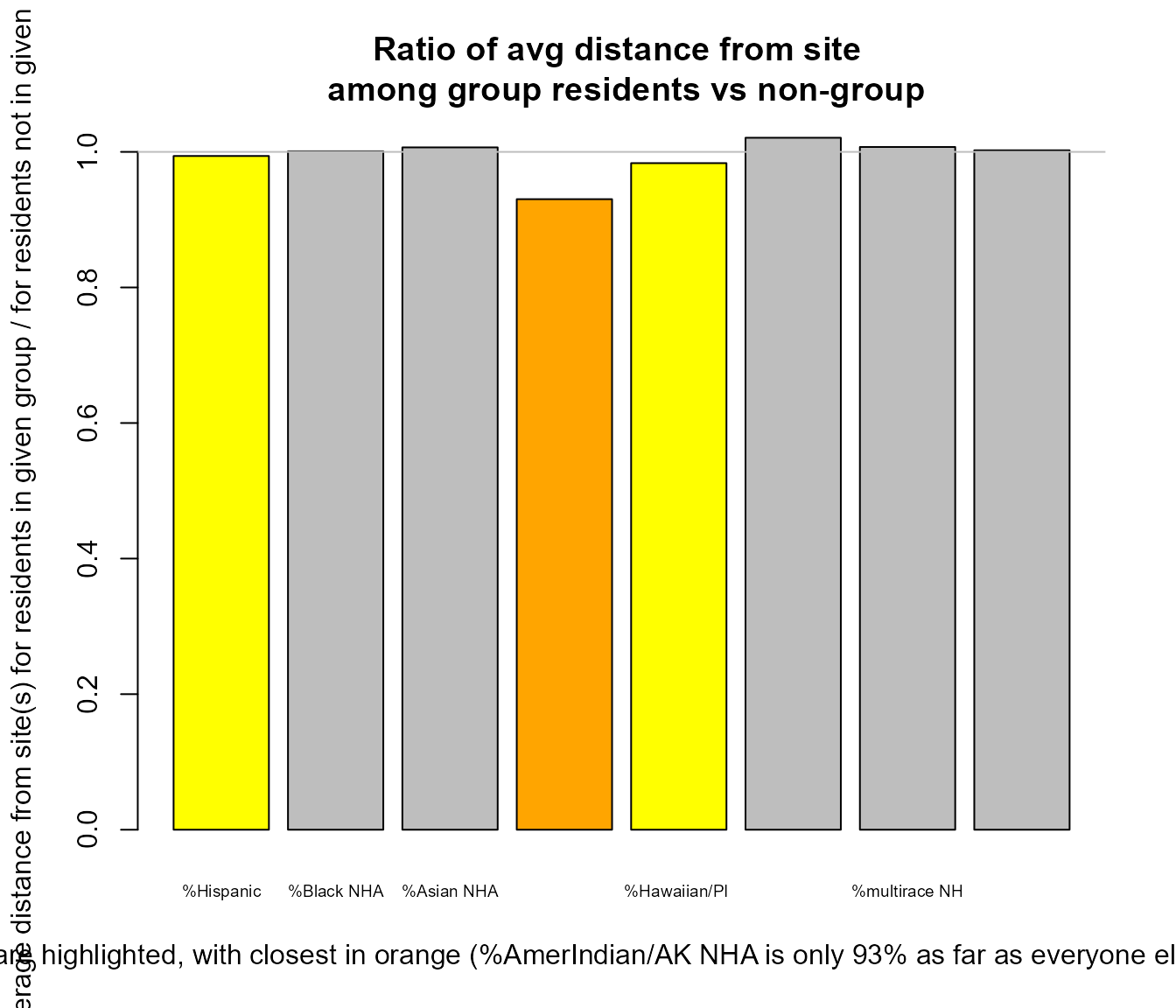
Example of using plot_distance_mean_by_group()
#> group nearest nearer ratio avg_distance_for_group
#> %Hispanic pcthisp FALSE TRUE 0.994 0.69
#> %Black NHA pctnhba FALSE FALSE 1.001 0.69
#> %Asian NHA pctnhaa FALSE FALSE 1.007 0.69
#> %AmerIndian/AK NHA pctnhaiana TRUE TRUE 0.930 0.64
#> %Hawaiian/PI NHA pctnhnhpia FALSE TRUE 0.983 0.68
#> %Other race NHA pctnhotheralone FALSE FALSE 1.021 0.70
#> %multirace NH pctnhmulti FALSE FALSE 1.007 0.69
#> %White NHA pctnhwa FALSE FALSE 1.002 0.69
#> avg_distance_for_nongroup
#> %Hispanic 0.69
#> %Black NHA 0.69
#> %Asian NHA 0.69
#> %AmerIndian/AK NHA 0.69
#> %Hawaiian/PI NHA 0.69
#> %Other race NHA 0.69
#> %multirace NH 0.69
#> %White NHA 0.69Site by site comparison
Mean distance of each group, at EACH SITE, as ratio to mean of everyone else nearby
Ratios at each site, of avg dist of group / avg dist of everyone else near site:
out <- testoutput_ejamit_10pts_1miles
## But try a larger radius to reveal more information:
# out <- ejamit(testpoints_10, radius = 31)
x = distance_by_group_by_site(out$results_bybg_people)
x
# summary of closest group at each site and by how much
data.frame(site = colnames(x),
closestgroup = rownames(x)[sapply(x, which.min)],
their_avg_distance_as_pct_of_everyone_elses = round(100 * sapply(x, min, na.rm = TRUE), 0)
)BACKGROUND AND OVERVIEW OF ISSUES IN PROXIMITY, DISTANCE, OR RADIUS
Distance from a potential source of environmental risk is often used as a simple proxy for actual exposure or risk, when data are limited. Proximity analysis uses distance (how far away) from a site, which is just the opposite of proximity (how near) to a site.
Conclusions can be sensitive to the choice of radius, if only one radius is reported on, as shown in [Step through all the sites to see an indicator versus distance at each].
Group’s percentage at each distance versus distance for each population group
Two basic ways to report residential population percentages and risk are 1) showing residential population percentage as a function or risk, and 2) showing risk as a function of residential population group:
Residential population group percentage as a function of risk (or proximity): Many proximity analyses report percentage by distance or risk bin, such as % low income within 3 miles of a point. This expresses residential population shares as a function of proximity or risk. Sometimes other distance or risk bins are used, such as areas with risk above some cutoff. And sometimes instead of a continuous measure of percentage, the residential population data are used to categorize places in bins, such as areas in the top quartile of poverty rates.
Risk (or proximity) as a function of residential population group: A different way to present this information is to report distance or risk as a function of residential population group – this expresses distance within each residential population group, such as the average distance by group or the full distribution of risk within each group.
Radius, radii, or continuous distance?
Proximity or distance as binary, categorical, or continuous metrics: Proximity analysis has often relied on picking a single distance, a radius, and analyzing conditions within that radius, such as all residents who live within 3 miles of a point where a regulated facility is located. Sometimes an analysis will look at two or even three distances. In some more sophisticated analyses, distance is treated as a continuous measure. Some tools like EJScreen use a proximity metric based on the inverse of distance (1/d) to provide a proximity score that gets higher as distance gets smaller. But many EJ analyses still use a single distance and analyze conditions within that distance.
EJAM makes it easier to do any of these types of analysis, because conclusions can be sensitive to the choice of a single radius, and metrics and methods provide different perspectives and reveal a richer picture of where people actually live in relation to potential sources of exposure or risk.
Comparisons within what distances or to what reference area(s)?
This is a tricky issue in proximity analysis: There is a subtle but vital difference between proximity analysis using a single radius (binary distance) and analysis using continuous distance. One way to think of this is that there are two aspects of or degrees of proximity to consider when analyzing residential population groups within a certain fixed distance (radius) from a single facility point (or a whole set of facilities). These two ways of summarizing proximity are complementary:
Which groups tend to live nearby in the sense of being within the radius versus outside the radius selected? In other words, which groups are “overrepresented” within X miles of the site? This treats proximity as a yes/no, binomial question – a resident is nearby or not. It would focus on whether someone is anywhere within 3 miles, say, and ignore the differences between being 1, 2, or 3 miles away. Most proximity analysis has tended to look at this type of summary.
Among the residents within X miles of the site, which groups live especially close to the facility? This question recognizes proximity is a continuous variable, and focuses on the difference between 1 mile, 1.5 miles, etc. However, it only looks at residents within the X miles radius area analyzed, so it fails to recognize that some groups tend to live more than 3 miles away, for example. This perspective does not take into account which groups are overrepresented within the original total radius near a site.
Some functions like distance_mean_by_group() or distance_by_group_by_site() do the second of these two types of analysis. They report, only among those anywhere inside the radius, which groups are closer to the site.
In a specific location, for example, one residential population group could be underrepresented within 3 miles, but those few who are in the group still might live right next to the facility in which case their average distance would be higher than that of any other group because this function only counts those within the radius analyzed.
In some other location, the opposite could occur – if one group is overrepresented within 3 miles, they still might all live in a community about 2.9 miles away from the site – that would mean their distance from the site on average is greater (or their proximity score is lower) than other groups within 3 miles of the site.
The question of whether to compare to Statewide or Nationwide or urban/rural or other reference averages or percentiles is related to this question of how to look at distances, or exposures or risk, just like it relates to how to look at residential population group percentages. One could look at percentage rate within 1 mile, 2 miles, etc. all the way out until one was looking at the county overall, the state overall, and eventually the nation overall. Selecting a single radius or selecting a single reference area should be done with a recognition of what questions one is actually trying to answer, and an understanding of how impacts vary with distance from a particular type of facility or source of potential risk.
If one is comparing residential population groups in terms of distance (or risk level), or if one is comparing % at each distance (or risk level), the implicit assumption is that there is some “expected” rate, and/or some “equitable” or “proportionate” % or ratio or risk.
CHOICE OF RADIUS AND UNCERTAINTY DUE TO A SMALL RADIUS WHERE BLOCKS ARE LARGE
Choosing a radius (or polygon) that is small relative to local Census blocks can lead to significant uncertainty in EJAM estimates, so it is important to understand the details if one wants to use a small radius especially in rural (low population density) areas.
To help consider this uncertainty, EJAM reports how many block centroids were found inside each area (inside a circular buffer defined by the selected radius, or inside a polygon that is from a shapefile). That count of blocks is found in a column of the spreadsheet output provided by the web app and also the table called results_bysite that is one output of the ejamit() function.
You could also Map all sites with popup at each saying how many blocks were found nearby and therefore might have more uncertainty in counts nearby.
# out <- ejamit(testpoints_1000, radius = 1)
# out$results_bysite$blockcount_near_site
out <- testoutput_ejamit_1000pts_1miles
barplot(
table(cut(
out$results_bysite$blockcount_near_site,
c(-1, 9, 29, 100, 1000)
)),
names.arg = c("< 10 blocks", "10-29", "30-100", "> 100 blocks"),
main = "How many blocks are within 1 mile of these 1,000 facilities?",
ylab = "# of facilities",
xlab = "# of blocks nearby"
)For more details about distance adjustments, overlaps of circles, etc.
This function prints a very large amount of diagnostic information, and provides a barplot histogram showing in this case that almost none of the 1000 sites have zero blocks within a mile but roughly 10-15% have under 10 blocks nearby and a similar share have only 10-29 blocks nearby.
# (Printed information is lengthy)
getblocks_diagnostics(
testoutput_getblocksnearby_1000pts_1miles,
# getblocksnearby(testpoints_1000, radius = 1, quiet = T),
detailed = T, see_pctiles = T
)Suggestions on radius and uncertainty
Here are some suggestions about how to consider the radius in relation to uncertainty where blocks are large:
- A closer look at uncertainty and care in communicating uncertainty may be needed where a circle or polygon contains fewer than about 30 block centroids. That is especially important if it contains fewer than about 10, and essential if it contains only 1 or zero block centroids.
- Using a radius of 5 miles or more does not raise these issues in 99% of US locations where EPA-regulated facilities are found.
- A radius of 3 miles might need a closer look for about 1% to 5% of typical sites in the US.
- A radius of 1 mile or less requires caution and understanding of the issues at a significant share of locations in the US (about 1 in 4 locations might need a closer look to check for uncertainties).
- A 0.5 mile radius should not be used without cautious interpretation or offline analysis in most locations where EPA-regulated facilities are located.
- A 0.25 mile radius should only be used on a case-by-case basis where each location is examined individually and other methods are likely more suited for the analysis of those sites.
These considerations are explained further in the discussion below.
Residential population group counts and percentages or environmental indicators are calculated from block group residential population data and environmental indicators and an estimate of what fraction of each block group is inside each site. For proximity analysis that means a circle is drawn around a point using a radius, and for shapefiles a similar approach is used. In either case, the fraction of the block group counted as inside the area analyzed is based on which block centroids (each is technically called a block “internal point”) are inside the circle or polygon. All the residents of a block are assumed to be inside if the block centroid is inside. This is exactly true unless a block is on the edge of the circle or polygon. Even for the ones on the edge, some centroids are just outside and some just inside the shape, so the contributions of some blocks are overcounted and other undercounted, but those tend to cancel each other out in the sense that it is unlikely they would all be undercounted, for example. Still, when a large share of the block points in circle or polygon are from blocks not entirely inside, uncertainty is higher than when the vast majority of blocks are entirely inside. In other words, if the circle or polygon has a very large number of blocks in it, uncertainty is lower because only a small fraction are along the edge and bisected. If a radius of 3 miles is used, the area is 28 square miles. If the blocks in that location are only about 0.28 square miles each, the circle might contain or partly contain about 100 blocks.
The dataset used by EJAM called blockwts has a column called block_radius_miles that is what the radius would be if the block were circular, and it was created based on area = pi * block_radius_miles^2 or block_radius_miles = sqrt(area / pi) where area is in square miles.
Details on the blocks found near one site
Table of distances between each site and each block
Use getblocksnearby() to quickly find residents/blocks
that are within a specified distance, as a table of distances between
sites and nearby blocks.
sitepoints <- testpoints_10[1:2, ]
sites2blocks <- getblocksnearby(
sitepoints = sitepoints,
radius = 3.1
)
#> Analyzing 2 points, radius of 3.1 miles around each.
#> Finding Census blocks with internal point within 3.1 miles of the site (point), for each of 2 sites (points)...
#> Stats via getblocks_diagnostics(), but NOT ADJUSTING UP FOR VERY SHORT DISTANCES:
#> min distance before adjustment: 0.06504053
#> max distance before adjustment: 4.670403
head(sites2blocks)
#> Key: <blockid>
#> ejam_uniq_id blockid distance blockwt bgid distance_unadjusted
#> <int> <int> <num> <num> <int> <num>
#> 1: 2 296 2.946818 0.00000000 15 2.946818
#> 2: 2 297 3.040476 0.00000000 15 3.040476
#> 3: 2 773 2.925869 0.07083906 32 2.925869
#> 4: 2 780 2.873082 0.03576341 32 2.873082
#> 5: 2 781 2.739460 0.05249885 32 2.739460
#> 6: 2 786 3.077032 0.03163686 32 3.077032Detailed stats on blocks found near site(s)
x <- getblocks_diagnostics(sites2blocks)
#>
#> DISTANCES FROM BLOCKS (AND RESIDENTS) TO SITES (AND FOR CLOSEST SITE)
#>
#> 3.097412 miles is max. distance to block internal point (distance_unadjusted)
#> 3.097412 miles is max. distance to average resident in block (distance reported)
#> 0.06504053 miles is shortest distance to block internal point (distance_unadjusted)
#> 0.06504053 miles is shortest distance to average resident in block (distance reported)
#> 0 block distances were adjusted (these stats may count some blocks twice if adjusted at 2+ sites)
#> 0 block distances were adjusted up (reported dist to avg resident is > dist to block internal point)
#> 0 block distances were adjusted down (reported < unadjusted)
#> 0 unique sites had one or more block distances adjusted due to large block and short distance to block point
#>
#> BLOCK COUNTS PER SITE (FEWER MEANS HIGHER UNCERTAINTY AT THOSE SITES)
#>
#> 362 blocks are near the avg site or in avg buffer
#> (based on their block internal point, like a centroid)
#>
#> sites blocks_per_site
#> 1 0 Not even 1
#> 2 0 some but <10
#> 3 0 10-29
#> 4 2 at least 30 blocks
#>
#> BLOCK COUNTS TOTAL AND IN OVERLAPS OF AREAS (MULTIPLE SITES FOR SOME RESIDENTS)
#>
#> 723 actual unique blocks total
#> 723 blocks including doublecounting in overlaps,
#> in final row count (block-to-site pairs table)
#> 1 is ratio of blocks including multicounting / actual count of unique blocks
#> 0% of unique blocks could get counted more than once
#> because those residents are near two or more sites
#> (assuming they live at the block internal point
#>
#> SITE COUNTS TOTAL AND IN OVERLAPS OF AREAS (MULTIPLE SITES FOR SOME RESIDENTS)
#>
#> 2 unique output sites
#>
#> 723 blocks (and their residents) have exactly 1 site nearby
#> 0 blocks (and their residents) have exactly 2 sites nearby
#> 0 blocks (and their residents) have exactly 3 sites nearby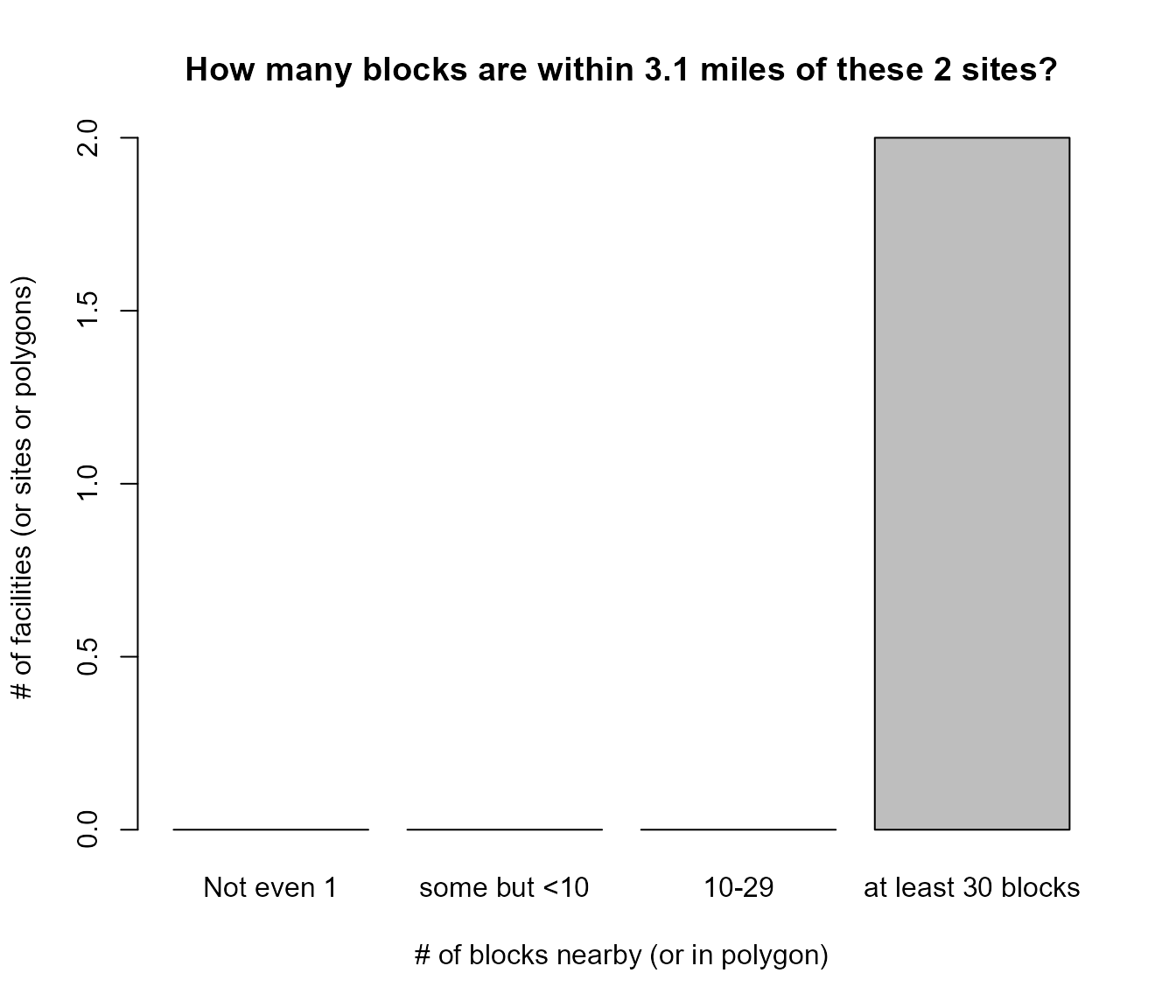
Example of getblocks_diagnostics() to see tables and histogram barplot of how many blocks are within 3.1 miles of these 2 sites
# x <- getblocks_summarize_blocks_per_site(sites2blocks)
# print(x) shows more info returned invisiblyMap 1 site to inspect the blocks nearby
Clicking on a block point provides a popup window showing information such as this:
blockfips: 131850102031056
blockid: 1788737
blocklat: 30.9913730000001
blocklon: -83.3753460999999
distance: 1.03614020347595
distance_unadjusted: 1.03614020347595
blockwt: 0
blockpop: 0
pop_nearby: 6237
bgpop: 1281
bgfips: 131850102031
bgid: 64286
ejam_uniq_id: 1
blockcount_near_site: 219
x <- plotblocksnearby(testpoints_10[1, ], radius = 3, returnmap = F)
#> Analyzing 1 points, radius of 3 miles around each.
#> Finding Census blocks with internal point within 3 miles of the site (point), for each of 1 sites (points)...
#> Stats via getblocks_diagnostics(), but NOT ADJUSTING UP FOR VERY SHORT DISTANCES:
#> min distance before adjustment: 0.2321264
#> max distance before adjustment: 4.412158
# Set returnmap= TRUE to actually return a leaflet mapPOPULATION DENSITY – WHY THE AVG SITE AND AVG RESIDENT ARE SO DIFFERENT
Reporting EJAM information summarized for the average site gives very different answers than reporting on the average resident near any one or more of those sites. The average site and average resident are completely different because most of the residents live near just a few of the sites – the ones with higher population density – when one is using a fixed radius at all sites, such as 3 miles from each site. Taking the average of sites gives equal weight to each site, even the ones with very few residents around them. Taking the average of all residents near all the sites gives equal weight to each person, so conditions near certain sites affect more people and have more influence on that average.
Sites vary widely in count of blocks nearby, depending on population density (which is closely related to block area in square miles)
- what blocks are near each site
- how far are they
- how many blocks are typically near a given site (population density varies)
- how many sites are near a block (residents with > 1 site nearby)
out <- testoutput_ejamit_100pts_1miles
cat(" ", popshare_p_lives_at_what_pct(out$results_bysite$pop, p = 0.50, astext = TRUE), "\n")
#> 6% of places account for 50% of the total population (approx.)
cat(" ", popshare_at_top_n(out$results_bysite$pop, c(1, 5, 10), astext = TRUE), "\n\n")
#> 1, 5, 10 places account for 17%, 46%, 58% of the total populationFind all blocks nearby each site
radius <- 3
sitepoints <- testpoints_100
sites2blocks <- getblocksnearby(sitepoints, radius, quadtree = localtree, quiet = TRUE)
#> Analyzing 100 points, radius of 3 miles around each.
# testoutput_getblocksnearby_10pts_1miles is also available as an example
names(sites2blocks)
#> [1] "ejam_uniq_id" "blockid" "distance"
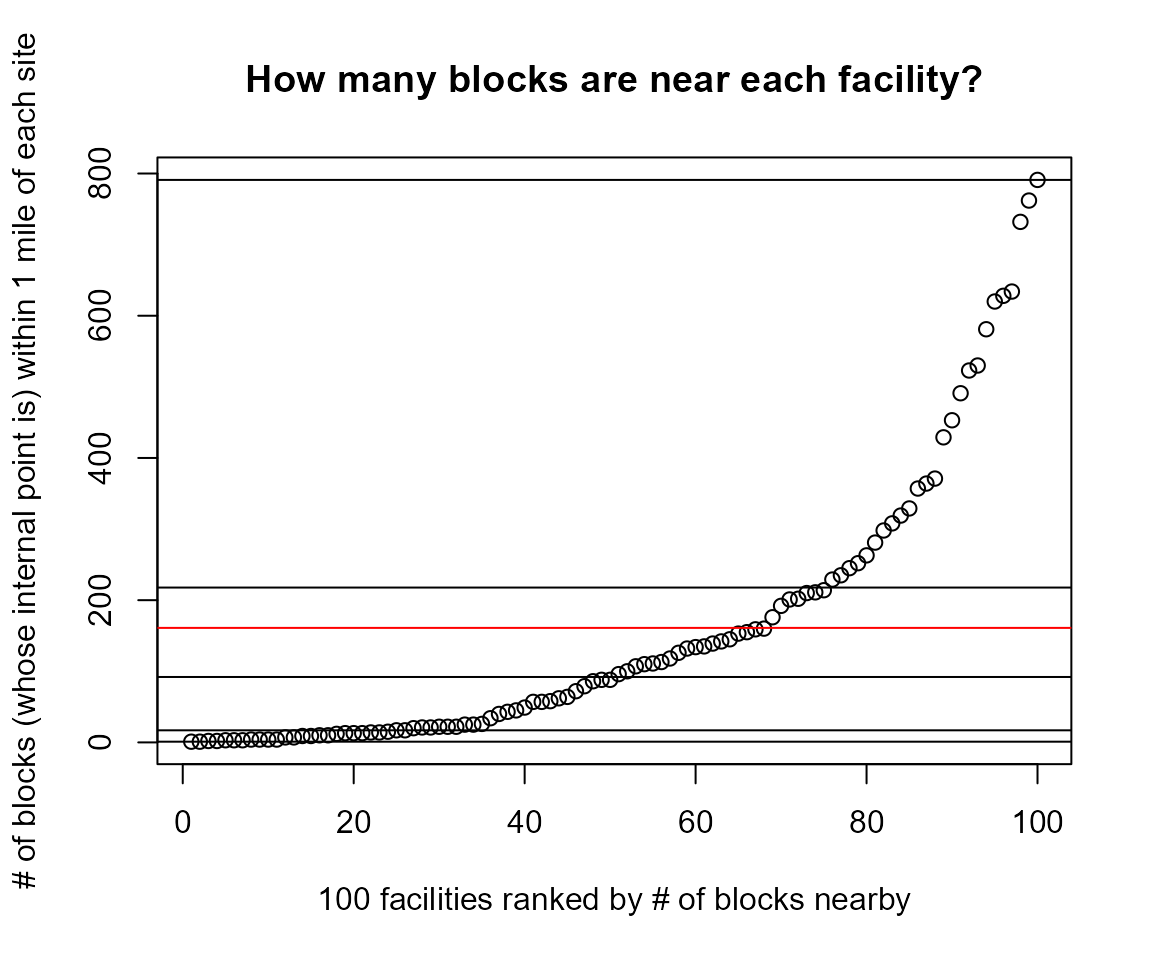
#> [4] "blockwt" "bgid" "distance_unadjusted"Very few blocks are within a radius of 1/4 mile.
Hundreds are often within 1 mile, but sometimes there are only a handful or even zero.
s2b_stats <- sites2blocks[ , .(
avgDistance = round(mean(distance), 2),
blocksfound = .N,
blocks_within_1mile = sum(distance <= 1),
blocks_within_0.75 = sum(distance <= 0.75),
blocks_within_0.25 = sum(distance <= 0.25)
), by = 'ejam_uniq_id'][order(blocksfound), ]
setorder(s2b_stats, ejam_uniq_id)
head(s2b_stats)
#> ejam_uniq_id avgDistance blocksfound blocks_within_1mile blocks_within_0.75
#> <int> <num> <int> <int> <int>
#> 1: 1 1.66 341 88 61
#> 2: 2 2.13 56 3 2
#> 3: 3 1.50 2111 620 378
#> 4: 4 1.65 735 210 130
#> 5: 5 0.90 405 298 255
#> 6: 6 2.00 48 4 4
#> blocks_within_0.25
#> <int>
#> 1: 6
#> 2: 0
#> 3: 36
#> 4: 12
#> 5: 50
#> 6: 0Histogram and table showing how many blocks are nearby a site
hist(sites2blocks[,.N, by = "ejam_uniq_id"][, N], 20,
xlab = "How many blocks are nearby?",
ylab = "Frequency (# of sites)",
main = "A given site may have zero to hundreds of blocks nearby",
sub = "A typical site in this example has about 100 blocks nearby")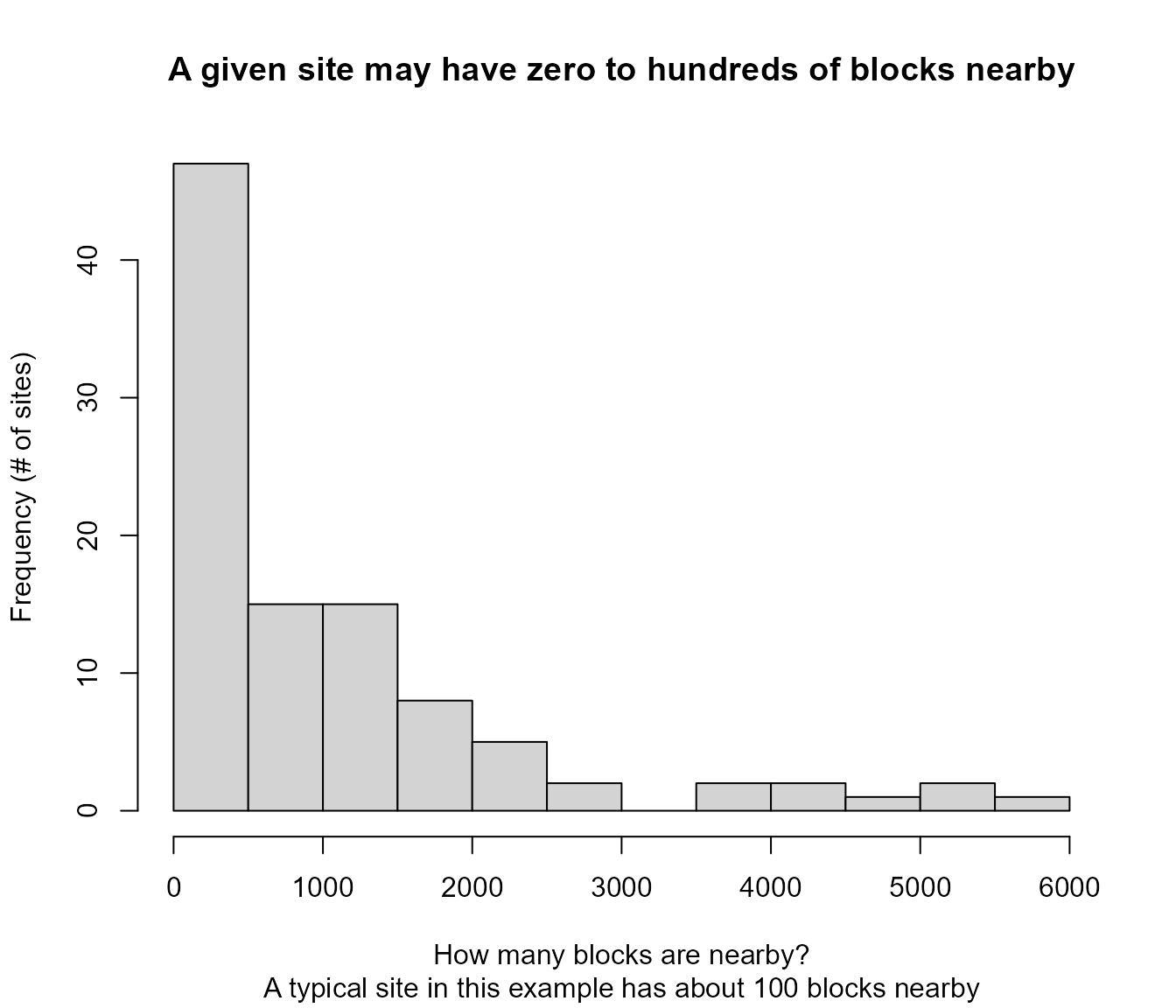
Example of Histogram and table showing how many blocks are within 3 miles of a site
DT::datatable(s2b_stats, rownames = FALSE)
# more summaries showing there may be only 1 block or hundreds within 1 mileMap all sites with popup at each saying how many blocks were found nearby
## done previously:
# radius <- 3
# sitepoints <- testpoints_100
out <- ejamit(sitepoints = sitepoints,
radius = radius, include_ejindexes = F)
#> Finding blocks nearby.
#> Analyzing 100 points, radius of 3 miles around each.
#> Aggregating at each site and overall.
#> Warning in batch.summarize(sitestats = data.frame(out$results_bysite), quiet =
#> quiet, : specified threshnames not all found in sitestats colnames, so using
#> defaults
few <- out$results_bysite$blockcount_near_site < 30
mapthis <- cbind(
sitepoints,
out$results_bysite[, c(
"pop", "bgcount_near_site", "blockcount_near_site"
)],
NOTE = ifelse(few, "< 30 blocks here", "")
)
# Show in red the sites with very few blocks nearby, suggesting more uncertainty in residential population group counts
mm <- mapfast(mapthis, radius = radius, color = 'navy')
mm |> leaflet::addCircles(
lng = mapthis$lon[few],
lat = mapthis$lat[few],
color = "red", radius = radius * 2 * meters_per_mile,
popup = popup_from_any(mapthis[few, ])
)Example of mapfast() for seeing how many blocks are at each site
Some places have hundreds nearby: a 1 mile radius is huge within a dense urban area
head(s2b_stats[order(s2b_stats$blocks_within_1mile, decreasing = T),
c('ejam_uniq_id', 'blocks_within_1mile')], 3)
#> ejam_uniq_id blocks_within_1mile
#> <int> <int>
#> 1: 57 791
#> 2: 42 762
#> 3: 21 732
densest <- s2b_stats$ejam_uniq_id[order(
s2b_stats$blocks_within_1mile, decreasing = T)][1]
leastdense <- s2b_stats$ejam_uniq_id[order(
s2b_stats$blocks_within_1mile, decreasing = F)][1]#> Analyzing 1 points, radius of 3 miles around each.
#> Finding Census blocks with internal point within 3 miles of the site (point), for each of 1 sites (points)...
#> Stats via getblocks_diagnostics(), but NOT ADJUSTING UP FOR VERY SHORT DISTANCES:
#> min distance before adjustment: 0.02342987
#> max distance before adjustment: 6.646285
plotblocksnearby(sitepoints = sitepoints[densest, ])#> Analyzing 1 points, radius of 3 miles around each.
#> Finding Census blocks with internal point within 3 miles of the site (point), for each of 1 sites (points)...
#> Stats via getblocks_diagnostics(), but NOT ADJUSTING UP FOR VERY SHORT DISTANCES:
#> min distance before adjustment: 0.5222611
#> max distance before adjustment: 5.281615
plotblocksnearby(sitepoints = sitepoints[ leastdense, ])Within a 1 mile radius, the blocks found tend to be about 2/3 of a mile from the site at the center.
summary(s2b_stats$avgDistance)
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> 0.900 1.765 1.930 1.853 2.033 2.500