Example Module 3 Workflow
TADA Team
2026-02-23
Source:vignettes/ExampleMod3Workflow.Rmd
ExampleMod3Workflow.RmdWelcome!
Thank you for your interest in Tools for Automated Data Analysis (TADA). TADA is an open-source tool set built in the R programming language. The EPATADA R Package is still under development. New functionality is added weekly, and sometimes we need to make bug fixes in response to user feedback. We appreciate your feedback, patience, and interest in these helpful tools. If you are interested in contributing to TADA development, more information is available here. We welcome collaboration with external partners!
Install and load packages
First, install and load the remotes package specifying the repo. This is needed before installing the EPATADA R package because it is only available on GitHub.
install.packages("remotes",
repos = "http://cran.us.r-project.org"
)
library(remotes)Next, install and load the EPATADA R Package using the remotes package. Dependency packages will also be downloaded automatically from CRAN. You may be prompted in the console to update dependencies. It is recommended to update all dependencies (enter 1 into the console).
remotes::install_github("USEPA/EPATADA",
ref = "develop",
dependencies = TRUE
)
# remotes::install_github("USGS-R/dataRetrieval", dependencies=TRUE)Finally, use the library() function to load the TADA R Package into your R session.
All EPATADA R package functions have their own individual help pages
(see Function
Reference). Users can also access function help pages from RStudio
by entering ?[name of TADA function] into the console
(example below).
# Access help page for TADA_DataRetrieval
?TADA_DataRetrievalModule 3 Functions in TADA
Disclaimer: The EPATADA Module 3 functions were designed to: (1) assist users with associating Water Quality Portal monitoring locations with assessment units and designated uses from ATTAINS and (2) compare Water Quality Portal results with numeric water quality criteria. EPATADA functions do not constitute current EPA policy or regulatory requirements. Organizations may choose to use EPATADA as a a tool in their decision making processes. Use of EPATADA is not required.
Introduction to TADA Module 3
This RMarkdown document walks users through how to create WQP and ATTAINS crosswalks that are needed to define and capture organization specific water quality analysis criteria and methodologies. This is a continuation of an example TADA workflow which began with ExampleMod2Workflow.Rmd.
Specifically, this vignette provides an overview of three functions that can assist users with:
Creating a crosswalk (reference table) between ATTAINS parameter names and WQP/TADA characteristic names/TADA comparable data identifier.
Creating a crosswalk (reference table) of all unique combinations of ATTAINS uses and parameters applicable to your analysis needs.
Define and summarize your spatial level of analysis for uses and parameters to locations(monitoring locations or assessment units) and capture any unique site-specific WQS criteria.
These functions are designed to be flexible so users from states, tribes, or territories can input organization-specific information and link it information from ATTAINS. While TADA functions can help generate these crosswalks and fill in some values, users must review and modify the tables generated in each step to ensure accuracy. The TADA team has also incorporated national recommended Clean Water Act (CWA) EPA 304(a) numeric criteria for optional use in Module 3 functions that has been filled out and reviewed by the TADA team. This allows users to analyze their data against the EPA 304(a) criteria; and to easily compare results when when using EPA 304(a) criteria vs. their own state or tribe’s criteria.
Getting Started
First, we will load example reference tables (crosswalks) created for TADA workflow example purposes. These are (1) a ML to AU crosswalk and (2) a Uses to AU crosswalk. If you would like to learn how these reference tables were created, see the ExampleMod2Workflow vignette.
# import example data set, example AUML crosswalk and example uses to AU crosswalk.
# extract ATTAINS_crosswalk data frame from the list
utils::data(Data_MT_AUMLRef)
Final.MT.AUMLRef <- Data_MT_AUMLRef$ATTAINS_crosswalk
utils::data(Data_MT_AU_UsesRef)
MT.AU_UsesRef <- Data_MT_AU_UsesRefGet WQP Monitoring Data in Montana Using TADA_DataRetrieval()
Let’s start with an example data frame that has already been through the data cleaning, wrangling, harmonization, handling of censored data, removal of suspect results, and other important TADA Module 1 functions. This process should be completed by users before utilizing Module 2 or 3 functions, so we will go through some minimal cleaning with an example data set from Montana. Get bacteria and pH data from Missoula County, Montana.
# get MT data
tada.MT <- TADA_DataRetrieval(
startDate = "2020-01-01",
endDate = "2022-12-31",
statecode = "MT",
characteristicName = c(
"Escherichia",
"Escherichia coli",
"pH"
),
countycode = "Missoula County",
ask = FALSE
)## [1] "Downloading WQP query results. This may take some time depending upon the query size."
## $statecode
## [1] "US:30"
##
## $startDate
## [1] "2020-01-01"
##
## $countycode
## [1] "Missoula County"
##
## $characteristicName
## [1] "Escherichia" "Escherichia coli" "pH"
##
## $endDate
## [1] "2022-12-31"
##
## [1] "Data successfully downloaded. Running TADA_AutoClean function."
## [1] "TADA_Autoclean: creating TADA-specific columns."
## [1] "TADA_Autoclean: handling special characters and coverting TADA.ResultMeasureValue and TADA.DetectionQuantitationLimitMeasure.MeasureValue value fields to numeric."
## [1] "TADA_Autoclean: converting TADA.LatitudeMeasure and TADA.LongitudeMeasure fields to numeric."
## [1] "TADA_Autoclean: harmonizing synonymous unit names (m and meters) to m."
## [1] "TADA_Autoclean: updating deprecated (i.e. retired) characteristic names."
## [1] "No deprecated characteristic names found in dataset."
## [1] "TADA_Autoclean: harmonizing result and depth units."
## [1] "TADA_Autoclean: creating TADA.ComparableDataIdentifier field for use when generating visualizations and analyses."
## [1] "NOTE: This version of the TADA package is designed to work with numeric data with media name: 'WATER'. TADA_AutoClean does not currently remove (filter) data with non-water media types. If desired, the user must make this specification on their own outside of package functions. Example: dplyr::filter(.data, TADA.ActivityMediaName == 'WATER')"
# clean up data set (minimal)
tada.MT.clean <- tada.MT |>
TADA_RunKeyFlagFunctions() |>
TADA_SimpleCensoredMethods() |>
TADA_HarmonizeSynonyms()## [1] "TADA_FlagFraction: Rows with Suspect sample fractions have been flagged but retained. Review these rows using the TADA.SampleFraction.Flag column before proceeding and/or set clean = TRUE."
## [1] "TADA_FlagSpeciation: Rows with Suspect speciations have been flagged but retained. Review these rows using the new TADA.MethodSpeciation.Flag column before proceeding and/or set clean = 'suspect_only' or 'both'."
## [1] "TADA_FlagMeasureQualifierCode: Dataframe does not include any information (all NAs) in MeasureQualifierCode."
## [1] "TADA_IDCensoredData: No censored data detected in your dataframe. Returning input dataframe with new column TADA.CensoredData.Flag set to Uncensored"
## [1] "Cannot apply simple censored methods to dataframe with no censored data results. Returning input dataframe."
## [1] "Warning: Your dataframe contains suspect metadata combinations in the following flag columns:"
## Flag_Column Result Count
## 1 TADA.SampleFraction.Flag 135
# remove intermediate objects
rm(tada.MT)
# if you cannot run TADA_DataRetrieval query, the example data set can be loaded by uncommenting the code below
# tada.MT.clean <- Data_MT_MissoulaCountyThis example will focus only on Montana. The remainder of this vignette will walk through how to fill out two crosswalk tables (TADA_ParametersForAnalysis and TADA_UsesForAnalysis) that are needed before we can start assigning applicable criteria and methodologies information for this specific ATTAINS organization (MTDEQ).
ATTAINS Domains and Allowable Values
A crosswalk between TADA characteristics and ATTAINS parameters is needed before we can integrate information from these two data sets. Before creating the TADA/WQP CharacteristicName and ATTAINS parameter crosswalk table (TADA_ParametersForAnalysis), let’s review all the unique TADA.ComparableDataIdentifiers (characteristic, fraction and speciation combinations) in the example data. How many results are available for each within the example data set?
# create table with counts of TADA.ComparableDataIdentifiers
TADA_FieldValuesTable(tada.MT.clean, field = "TADA.ComparableDataIdentifier")## Value Count
## 1 PH_NONE_NONE_NONE 280
## 2 ESCHERICHIA COLI_NA_NA_CFU/100ML 146In this vignette, we will crosswalk each of these TADA.ComparableDataIdentifiers to ATTAINS parameter names. ATTAINS has multiple domains, including the parameter names domain, that have allowable values. Using the R package rExpertQuery, which provides functions for downloading tidy data from the ATTAINS public web services (https://github.com/USEPA/rExpertQuery), we can review all ATTAINS domains (see example below).
# return ATTAINS parameter domain values
TADA_TableExport(rExpertQuery::EQ_DomainValues("param_name"))## [1] "EQ_DomainValues: For param_name the values in the 'name' column of the function output are the allowable values for rExpert Query functions."In the next section, we will review which parameters have been listed in ATTAINS in the past for a specific organization. In order to select a specific organization in the TADA_ParametersForAnalysis() function, we need to identify the organization id used in ATTAINS. The organization id for Montana is “MTDEQ”.
# return ATTAINS organization domain values
TADA_TableExport(rExpertQuery::EQ_DomainValues("org_id"))## [1] "EQ_DomainValues: For org_id the values in the 'code' column of the function output are the allowable values for rExpert Query functions."TADA_ParametersForAnalysis() Basics
ATTAINS Parameter Names are more general than the TADA Comparable Data Identifiers. Therefore, we recommend users provide a crosswalk between each TADA.ComparableDataIdentifier and ATTAINS.ParameterName. TADA_ParametersForAnalysis() creates a template which includes all TADA.ComparableDataIdentifiers from the TADA data frame and provides a blank column, “ATTAINS.ParameterName” for users to input the corresponding ATTAINS parameter name.
TADA_ParametersForAnalysis() includes an argument input ‘auto_assign’ to assist users in finding an alias name between TADA Characteristic Names to ATTAINS Parameter Name, if one is found. It is important to note that even with these exact matches, there are additional complexities related to fraction, speciation and units that may be unique to your organization. For example, some organizations may consider Total Nitrogen, Nitrate/Nitrite, and Ammonia all to match the ATTAINS parameter “Nitrogen”, while others may consider only some fraction/speciation combinations analogous to the ATTAINS parameter “Nitrogen”.
Setting “excel = TRUE” in TADA_ParametersForAnalysis tells the function to create an Excel spreadsheet of the reference table for review and editing. The excel output includes logic to help a user select ATTAINS parameter names that have been used in the past by the selected organization. If users do not want to work in Excel, they can use “excel = FALSE” to return a data frame. The data frame can be edited directly in R if desired, although there are no TADA-specific functions designed to facilitate this.
When using TADA_ParametersForAnalysis(), users should specify the organization(s) of interest in the “org_id” argument. This ensures that the correct number of rows will be created in the reference table. When “excel = TRUE”, specifying the organization(s) also creates a separate tab in the Excel file which contains the parameter names used in prior assessment cycles by the specified ATTAINS organization(s). This will allow users to decide whether to continue to use the same ATTAINS parameter names from prior assessments that their organizations(s) have used in the past, or to use other valid parameter names from the entire ATTAINS domain list.
Keep in mind that only parameter names previously entered in ATTAINS by the selected organization(s) will be returned. Many organization only input parameters that are causes in ATTAINS. This means that TADA_ParametersForAnalysis() may not return all parameters required by an organization’s assessment methodology if some parameters have never been listed as a cause. If there is not a suitable ATTAINS parameter name to match a TADA.ComparableDataIdentifier, users should contact the ATTAINS team (attains@epa.gov) for assistance.
For our first example, we will specify Montana’s organization identifier (“MTDEQ”) as our ATTAINS organization identifier of interest. By default, auto_assign = “None” which will not populate any entries for the ATTAINS.ParameterName column, and users will need to manually fill this crosswalk out.
# create TADA parameter reference table for specified organization
MT.ParamRef.None <- TADA_ParametersForAnalysis(
tada.MT.clean,
org_id = c("MTDEQ"),
auto_assign = "None",
excel = FALSE
# uncomment to run the excel file
# excel = TRUE, overwrite = TRUE
)
TADA_TableExport(MT.ParamRef.None)Auto_assign Options
Now, let’s see what happens we we use auto_assign = ‘All’ to autopopulate exact matches. The auto assignment will create exact matches based on the entire list of allowable ATTAINS parameters. This means that parameter names your organization has not previously record in ATTAINS may be included in the assignments.
# create TADA parameter reference table for specified organization
MT.ParamRef.All <- TADA_ParametersForAnalysis(
tada.MT.clean,
org_id = c("MTDEQ"),
auto_assign = "All",
excel = FALSE
# excel = TRUE, overwrite = TRUE
)## [1] "TADA_ParametersForAnalysis: auto_assign == 'All' was selected, finding an exact ATTAINS.ParameterName match for each TADA.ComparableDataIdentifier - by WQP CharacteristicName if one is found."
TADA_TableExport(MT.ParamRef.All)Another option is to set auto_assign = ‘Org’. This limits the exact matches returned to only ATTAINS.ParameterNames the selected organization has used in the past.
# create TADA parameter reference table for specified organization
MT.ParamRef.Org <- TADA_ParametersForAnalysis(
tada.MT.clean,
org_id = c("MTDEQ"),
auto_assign = "Org",
excel = FALSE
# uncomment excel = TRUE, overwrite = TRUE to run the excel file
# excel = TRUE, overwrite = TRUE
)## [1] "TADA_ParametersForAnalysis: auto_assign == 'Org' was selected, finding an exact ATTAINS.ParameterName match, by ATTAINS.OrganizationName, for each TADA.ComparableDataIdentifier - by WQP CharacteristicName if one is found."
TADA_TableExport(MT.ParamRef.Org)Manual Parameter Crosswalk
The code chunk below demonstrates how the parameter reference table can be modified in R. Once modifications are complete, we can re-run TADA_ParametersForAnalysis using the modified parameter reference table as the input for “paramRef”. This will update the ATTAINS.FlagParameterName column in the parameter reference table (see example below). This column provides information about whether or not the organization specified in the “ATTAINS.OrganizationIdentifier” column has listed this parameter as a cause in prior assessment cycles. It also flags rows where the TADA.ComparableDataIdentifier as not been assigned an ATTAINS parameter name.
# only run the code chunks below if you make edits to the excel file. Please note you must open and save the excel file at least once to reflect all Excel formula based values.
# downloads_path <- file.path(Sys.getenv("USERPROFILE"), "Downloads", "myfileRef.xlsx")
# ParamRef <- openxlsx::read.xlsx(downloads_path, sheet = "CreateParamRef")
# MT.ParamRef_Excel <- TADA_ParametersForAnalysis(
# Data_NCTC,
# org_names = c("MTDEQ"),
# paramRef = ParamRef,
# excel = TRUE, overwrite = TRUE
# )
ParamRef <- MT.ParamRef.None |>
dplyr::mutate(ATTAINS.ParameterName = dplyr::case_when(
grepl("PH_NONE_NONE_NONE", TADA.ComparableDataIdentifier) ~ "PH",
grepl("ESCHERICHIA COLI", TADA.ComparableDataIdentifier) ~ "ESCHERICHIA COLI (E. COLI)"
)) |>
dplyr::bind_rows(data.frame(TADA.ComparableDataIdentifier = "PH_NONE_NONE_NONE", ATTAINS.ParameterName = "PH, HIGH", ATTAINS.OrganizationIdentifier = "MTDEQ"))
MT.ParamRef.Manual <- TADA_ParametersForAnalysis(
tada.MT.clean,
org_id = c("MTDEQ"),
paramRef = ParamRef,
excel = FALSE
# uncomment excel = TRUE, overwrite = TRUE to run the excel file
# excel = TRUE, overwrite = TRUE
)
TADA_TableExport(MT.ParamRef.Manual)Provide a User Supplied paramRef
We can now save and reuse MT.ParamRef_Manual as the user-supplied paramRef argument input in TADA_ParametersForAnalysis(). When this reference file is used for the next assessment or analysis cycle, this function will determine if any new WQX characteristic names are included in the TADA data frame and add additional rows to the crosswalk if needed.
MT.ParamRef.user.supplied <- TADA_ParametersForAnalysis(
tada.MT.clean,
org_id = c("MTDEQ"),
paramRef = MT.ParamRef.Manual,
excel = FALSE
# uncomment excel = TRUE, overwrite = TRUE to run the excel file
# excel = TRUE, overwrite = TRUE
)
TADA_TableExport(MT.ParamRef.user.supplied)Review, Save and Re-Use paramRef
Once the parameter reference table has been reviewed and modified, it can be saved and reused for analysis of future TADA data frames. This process is useful to help identify if new WQP characteristics (or new fraction/speciations) are being queried in your most up to date WQP query and to allow you to review how to handle these new additions.
If new WQP characteristics (or new fraction/speciations) do show up, your user supplied ‘paramRef’ is prioritized over the ‘auto_assign’ argument inputs. The auto_assign option will allow you to fill in any remaining blank ATTAINS.ParameterName that you have not filled in but will not replace any cross walk that you have defined in your user supplied crosswalk. In our case, since each unique TADA.ComparableDataIdentifier was cross walked to an ATTAINS.ParameterName in the user supplied ’MT.ParamRef_user_supplied, this table will be the same.
MT.ParamRef.Final <- TADA_ParametersForAnalysis(
tada.MT.clean,
org_id = c("MTDEQ"),
paramRef = MT.ParamRef.user.supplied,
auto_assign = "All",
excel = FALSE
# uncomment excel = TRUE, overwrite = TRUE to run the excel file
# excel = TRUE, overwrite = TRUE
)## [1] "TADA_ParametersForAnalysis: auto_assign == 'All' was selected, finding an exact ATTAINS.ParameterName match for each TADA.ComparableDataIdentifier - by WQP CharacteristicName if one is found."
# Test if the two data frames are same or not.
identical(MT.ParamRef.Final[1:4], MT.ParamRef.user.supplied[1:4])## [1] TRUE
TADA_TableExport(MT.ParamRef.Final)Remove intermediate variable, we will keep only the crosswalk tables that are relevant for the remaining workflow.
rm(ParamRef, MT.ParamRef.All, MT.ParamRef.Manual, MT.ParamRef.None, MT.ParamRef.Org, MT.ParamRef.user.supplied)TADA_UsesForAnalysis() Basics
Our first example will pull in all prior ATTAINS uses (ATTAINS.UseNames) and parameter names (ATTAINS.ParameterName) from the specified ATTAINS organization (defined by the org_id function argument for TADA_UsesForAnalysis), which in this case is “MTDEQ” (Montana) using ExpertQuery web services. Users will review the output and choose which ATTAINS.UseName(s) are applicable to their analysis. Later in the data analysis process, users will be asked to define criteria and methodologies that is applicable to each ATTAINS use name and parameter name labeled as “Include”. If an ATTAINS.UseName is not applicable, users should choose “Exclude” for that ATTAINS parameter name and use name.
Manual Uses to Parameter Assignment
Any ATTAINS parameter name(s) that have not been used by an organization in prior assessment cycles, will not have any associated prior use names. In this case, users must manually assign the appropriate ATTAINS.UseName under the column ‘ATTAINS.UseName’. They may need to add additional rows if the parameter applies to multiples uses. Users can also choose to ‘auto_assign’ all unique ATTAINS.UseName by ATTAINS.OrganizationName to any ATTAINS.ParameterName missing a use name associated with it.
In the example below, we can see “PH” and “ESCHERICHIA COLI (E. COLI)” as the ATTAINS.ParameterNames that were assessed in prior assessment cycles for MTDEQ (see ATTAINS.FlagUseName column in NCTC_usesRef). “PH, HIGH” was not listed in ATTAINS for MTDEQ in the prior assessment cycle, but was included as a parameter name MTDEQ would like to assess for in this current assessment cycle. Thus, the ATTAINS.UseName is left blank for “PH, HIGH”.
MT.usesRef.Manual <- TADA_UsesForAnalysis(
tada.MT.clean,
org_id = c("MTDEQ"),
paramRef = MT.ParamRef.Final,
auto_assign = FALSE,
excel = FALSE
# uncomment excel = TRUE, overwrite = TRUE to run the excel file
# excel = TRUE, overwrite = TRUE
)
TADA_TableExport(MT.usesRef.Manual)If desired, a user can manually assign “PH, HIGH” to any applicable uses (disclaimer: this is for demonstration purposes only and does not reflect MTDEQ’s criteria and assessment process).
add.data <- data.frame(
"ATTAINS.OrganizationIdentifier" = "MTDEQ",
"ATTAINS.ParameterName" = rep("PH, HIGH", 2),
"ATTAINS.UseName" = c(
"Aquatic Life",
"Agriculture"
)
)The output of this will not reflect changes to the ATTAINS.FlagUseName column. To do so, we need to re run TADA_UsesForAnalysis() with usesRef = add_data as an argument input.
usesRef <- MT.usesRef.Manual |>
dplyr::left_join(add.data, by = c("ATTAINS.OrganizationIdentifier", "ATTAINS.ParameterName"), keep = FALSE) |>
dplyr::mutate(ATTAINS.UseName = dplyr::coalesce(ATTAINS.UseName.x, ATTAINS.UseName.y)) |>
dplyr::select(-c(ATTAINS.UseName.x, ATTAINS.UseName.y)) |>
dplyr::mutate(IncludeOrExclude = "Include")
# PH will now reflect the changes
MT.usesRef.Manual.Update <- TADA_UsesForAnalysis(
tada.MT.clean,
paramRef = MT.ParamRef.Final,
usesRef = usesRef, # Edits were made to usesRef, updates flag column
org_id = c("MTDEQ"),
auto_assign = FALSE,
excel = FALSE
# uncomment excel = TRUE, overwrite = TRUE to run the excel file
# excel = TRUE, overwrite = TRUE
)
TADA_TableExport(MT.usesRef.Manual.Update)Auto_assign Option
Alternatively, we can choose to assign all unique use names found in your current usesRef table, by your organization to those ATTAINS.ParameterName without any associated ATTAINS.UseName. Users will need to review this assignment carefully, choose include or exclude appropriately, and include additional rows as needed if there are more ATTAINS.UseName applicable for that ATTAINS.ParameterName that was not captured by the auto_assign method.
# uses will come from the user supplied reference table produced in ExampleMod2Workflow.Rmd
utils::data(Data_MT_AU_UsesRef_Water)
MT.usesRef.AutoAssign <- TADA_UsesForAnalysis(
tada.MT.clean,
org_id = c("MTDEQ"),
paramRef = MT.ParamRef.Final,
auto_assign = TRUE,
excel = FALSE
# uncomment excel = TRUE, overwrite = TRUE to run the excel file
# excel = TRUE, overwrite = TRUE
)## [1] "TADA_UsesForAnalysis: auto_assign == TRUE was selected, assigning all unique ATTAINS.UseName, by ATTAINS.OrganizationIdentifier, to any ATTAINS.ParameterName that an organization have not done assessments for in prior ATTAINS cycle. Please review carefully and Exclude rows as needed."
TADA_TableExport(MT.usesRef.AutoAssign)User has completed a Use to AU crosswalk
Now, if a user has completed assignments of uses to AUs in the recommended ExampleMod2Workflow.Rmd, they need to provide this as a AU_UsesRef argument input. This will help to ensure that the uses are applicable to your most recent assessment/analysis needs. These uses are extracted based on the uses to water type assignments in your AU_UsesRef and joined to an internal parameter and water type extraction from rExpertQuery to assign any new uses to parameters.
MT.usesRef.with.AU_UsesRef <- TADA_UsesForAnalysis(
tada.MT.clean,
org_id = c("MTDEQ"),
paramRef = MT.ParamRef.Final,
AU_UsesRef = Data_MT_AU_UsesRef_Water, # uses will come from the user supplied reference table produced in ExampleMod2Workflow.Rmd
auto_assign = TRUE,
excel = FALSE
# excel = TRUE, overwrite = TRUE
)## [1] "TADA_UsesForAnalysis: auto_assign == TRUE was selected, assigning all unique ATTAINS.UseName, by ATTAINS.OrganizationIdentifier, to any ATTAINS.ParameterName that an organization have not done assessments for in prior ATTAINS cycle. Please review carefully and Exclude rows as needed."Provide a User Supplied usesRef
Users can also choose to supply their own usesRef as an argument input. This may be useful if your organization has made significant changes to use names in the current cycle (cannot be retrieved from the prior ATTAINS assessment cycle) or if there are just additional use names that were unable to be retrieved from the prior assessment cycle.
(Note: TADA functions leverages the ATTAINS assessment profiles which will not reflect any submitted use name changes to ATTAINS until the next assessment cycle is approved. In addition, it may not be possible to retrieve every use and parameter name combination from ATTAINS as it is dependent on what information each organization has submitted to ATTAINS.)
In this example, let’s use a filled out criteria table for MTDEQ and extract the use and parameters from their crosswalk.
We can see that MTDEQ has additional rows for E. Coli that was unable to be retrieved from ATTAINS. We also see PH was not found in the user supplied usesRef table. We will assume MTDEQ has forgotten to include PH in their table and showcase the importance of considering all readily available data from your WQP data retrieval that you may need to consider for analysis.
# Load the example MTDEQ criteria table
criteria_table <- system.file("extdata", "criteria_table.rda", package = "EPATADA")
load(criteria_table)
MT.UseParam.user.supplied <- dplyr::select(criteria_table, ATTAINS.OrganizationIdentifier, TADA.ComparableDataIdentifier, TADA.CharacteristicName, ATTAINS.ParameterName, ATTAINS.UseName)
MT.usesRef.user.supplied.edit <- TADA_UsesForAnalysis(
tada.MT.clean,
org_id = c("MTDEQ"),
paramRef = MT.ParamRef.Final,
usesRef = MT.UseParam.user.supplied,
auto_assign = TRUE,
excel = FALSE
# uncomment excel = TRUE, overwrite = TRUE to run the excel file
# excel = TRUE, overwrite = TRUE
)## [1] "TADA_UsesForAnalysis: auto_assign == TRUE was selected, assigning all unique ATTAINS.UseName, by ATTAINS.OrganizationIdentifier, to any ATTAINS.ParameterName that an organization have not done assessments for in prior ATTAINS cycle. Please review carefully and Exclude rows as needed."
## [1] "IncludeOrExclude was not found as a column name in your user supplied, assuming all parameter and uses are applicable for your analysis."
TADA_TableExport(MT.usesRef.user.supplied.edit)Review, Save and Re-Use usesRef
Once the uses to parameter reference table has been reviewed and modified, it can be saved and reused for analysis of future TADA data frames. This process is useful to help identify if any new WQP characteristics (or new fraction/speciations) are being queried in your most up to date WQP query and to allow you to review how to handle assigning ATTAINS uses to these new additions.
If new WQP characteristics (or new fraction/speciations) do show up, your user supplied ‘usesRef’ is prioritized over the ‘auto_assign’ argument inputs. Thus, the auto_assign option will allow you to fill in any remaining blank ATTAINS.UseName to an ATTAINS.ParameterName that you have not filled in but will not replace any cross walk that you have defined in your user supplied crosswalk.
MT.usesRef.Final <- TADA_UsesForAnalysis(
tada.MT.clean,
org_id = c("MTDEQ"),
paramRef = MT.ParamRef.Final,
usesRef = MT.usesRef.user.supplied.edit,
auto_assign = "All",
excel = FALSE
# uncomment excel = TRUE, overwrite = TRUE to run the excel file
# excel = TRUE, overwrite = TRUE
)
# Test if the two data frames are same or not.
identical(MT.usesRef.Final[1:5], MT.usesRef.user.supplied.edit[1:5])## [1] TRUE
TADA_TableExport(MT.usesRef.Final)remove intermediate variable, keep only the crosswalk table that we will continue to use in the workflow.
rm(MT.usesRef.AutoAssign, MT.UseParam.user.supplied, MT.usesRef.Manual, MT.usesRef.Manual.Update)TADA_MLSummary(): Create and Define Spatial Reference Tables
We can define your WQP data set on either an assessment unit level summary or monitoring location sites level. Please refer to EPATADA Module 2 for more information on the geospatial functions to assist with assessment unit and monitoring location overlay.
The general workflow of running module 3 functions for CWA water quality data analysis is shown below.
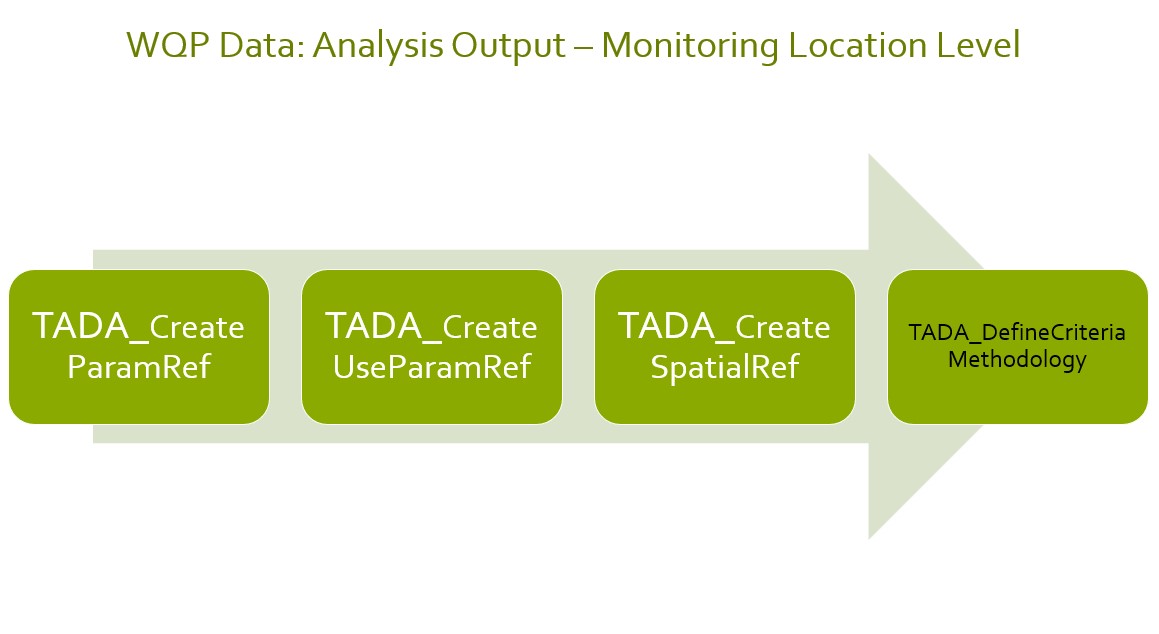
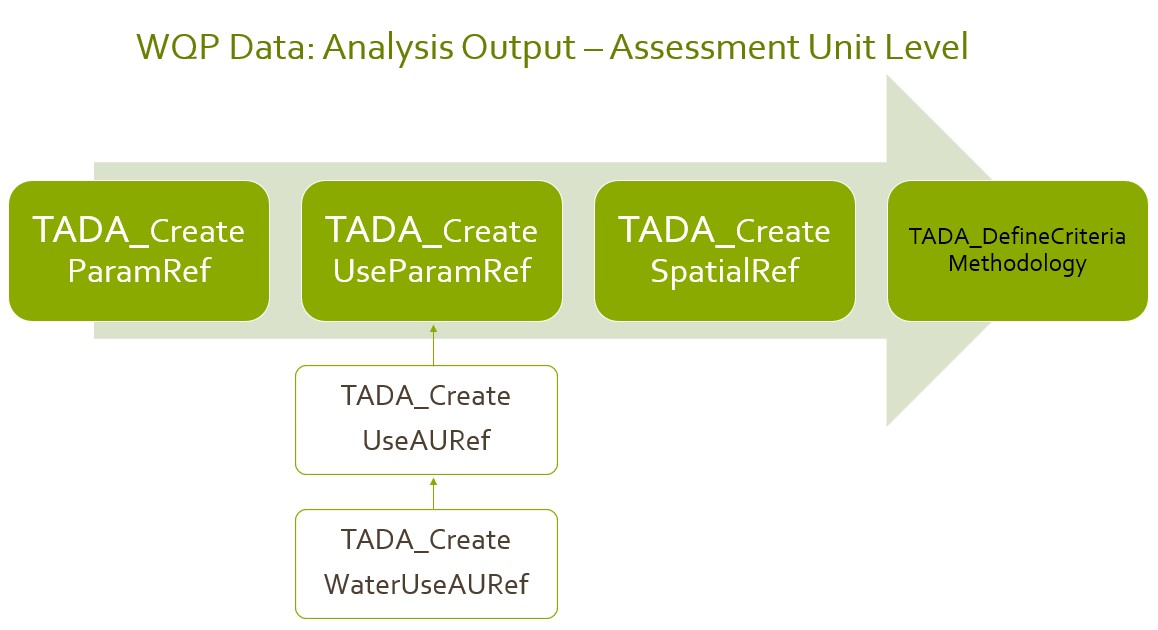
Define Spatial Summary by Monitoring Location (ML)
Let’s start off with a ML summary of MTDEQ data frame. By default, we will only display rows for parameters and uses for a ML if it contains data collected for that TADA.CharacteristicName in your WQP data query. If you would like to display all information even for sites with no WQP data, please choose displayNA = TRUE
MT.MLSummaryRef.ML <- TADA_MLSummary(
.data = tada.MT.clean,
usesRef = MT.usesRef.Final,
org_id = "MTDEQ",
displayNA = TRUE,
excel = FALSE
# excel = TRUE, overwrite = TRUE
)## [1] "TADA_MLSummary: displayNA = TRUE: This MLSummaryRef table will display ALL parameters and uses for a ML/AU regardless if it contains data collected for that TADA.CharacteristicName in your TADA data frame."
TADA_TableExport(MT.MLSummaryRef.ML)Define Spatial Summary by Monitoring Location (AU)
Now let’s compare this to the AU level of summary. To provide the ML to AU level of summary, this requires an AUML crosswalk and an AU_UsesRef, we recommend you to review the ExampleMod2Workflow vignette for more in depth information on this crosswalk. We will load these example reference tables into this vignette.
We now have all assignments of ATTAINS Uses, ATTAINS Parameters, and WQP Monitoring Location Sites to each AU defined. Users can do a final review to ensure all assignments are correct.
Note: By default, we will only display rows for parameters and uses for a ML if it contains data collected for that TADA.CharacteristicName in your WQP data query. If you would like to display all information even for sites with no WQP data, please choose displayNA = TRUE
# Load the dataset
utils::data("Data_MT_AU_UsesRef_Water", package = "EPATADA")
MT.MLSummaryRef.AU <- TADA_MLSummary(
.data = tada.MT.clean,
usesRef = MT.usesRef.Final,
AU_UsesRef = Data_MT_AU_UsesRef_Water, # uses will come from the user supplied reference table produced in ExampleMod2Workflow.Rmd
AUMLRef = Final.MT.AUMLRef,
org_id = "MTDEQ",
displayNA = FALSE,
excel = FALSE
# excel = TRUE, overwrite = TRUE
)
TADA_TableExport(MT.MLSummaryRef.AU)Assign site-specific spatial criteria
We can apply any unique spatial criteria on a monitoring location sites level. This view allows us to determine whether there are certain sites that needs a unique site-specific criteria, certain water types, or certain combination of characteristics that have site-specific criteria. Let’s go through an example of how a user may modify this table.
MT.MLSummaryRef.AU2 <- MT.MLSummaryRef.AU |>
dplyr::mutate(
UniqueSpatialCriteria = dplyr::case_when(
MonitoringLocationIdentifier == "MTVOLWQM_WQX-CLEARWATERR_1" ~ "Example Site Specific"
)
)
TADA_TableExport(MT.MLSummaryRef.AU2)Compare the nrow for each of the ML versus AU level of summary. The AU level of summary is further filtered down by the AU to ML crosswalk, the use to AU crosswalk, and the use to parameter crosswalk based on what TADA.ComparableDataIdentifier has been collected at those sites and taking into consideration of what parameter(s) and use(s) have been assessed for each assessment unit. (Note: Would users find it useful to still include a parameter and use for a site even if the site does not contain that TADA.ComparableDataIdentifier - the ATTAINS.ParameterName? This could be included in the final summary table and labeled as NA or insufficient data.)
TADA DefineCriteriaMethodology()
Now, lets get to generating the criteria and methodology file for MTDEQ to fill out. We will showcase how this table will be generated for the first time using the recommended step-by-step workflow and showcase how a user can go about updating their criteria and methodology file as needed.
First, let’s show how our step-by-step process all come together. Users will need to fill out this template that is generated. It is highly recommended to export this to the excel spreadsheet to show the allowable values and easy interface for inputs to the table.
MT.CriteriaMethods <- TADA_DefineCriteriaMethodology(
.data = tada.MT.clean,
org_id = "MTDEQ",
MLSummaryRef = MT.MLSummaryRef.ML,
excel = FALSE
# excel = TRUE, overwrite = TRUE
)## [1] "TADA_DefineCriteriaMethodology: displayUniqueId == FALSE was selected, TADA.ComparableDataIdentifier is converted to NA and duplicated rows are removed. Users are recommended to fill out any applicable combinations of Characteristic, Fraction and Speciation for analysis."
TADA_TableExport(MT.CriteriaMethods)Now, if MTDEQ, already has a filled out/partially filled out criteria methods table, let’s provide this as an argument input. criteria_table is an example of a criteria table that has been filled out by a few R8 states and tribes.
Note: Whenever a criteriaMethods argument input value is provided, users will be warned if there are additional WQP characteristics (or TADA.ComparableDataIdentifier - see argument input uniqueDataId = TRUE/FALSE in the R documentation for more information) that are not captured in their user supplied table. Thus, auto_assign defaults to TRUE even if it is specified as FALSE.
MT.CriteriaMethods.user.supplied <- TADA_DefineCriteriaMethodology(
.data = tada.MT.clean,
org_id = "MTDEQ",
MLSummaryRef = NULL,
criteriaMethods = criteria_table,
excel = FALSE
# excel = TRUE, overwrite = TRUE
)## [1] "TADA_DefineCriteriaMethodology: displayUniqueId == FALSE was selected, TADA.ComparableDataIdentifier is converted to NA and duplicated rows are removed. Users are recommended to fill out any applicable combinations of Characteristic, Fraction and Speciation for analysis."
TADA_TableExport(MT.CriteriaMethods.user.supplied)You can append EPA304(a) recommended standards by specifying “USEPA” as part of org_id (if there is one found for a TADA.CharacteristicName)
MT.CriteriaMethods.user.supplied2 <- TADA_DefineCriteriaMethodology(
.data = tada.MT.clean,
org_id = c("MTDEQ", "USEPA"),
MLSummaryRef = NULL,
criteriaMethods = criteria_table,
displayUniqueId = TRUE,
excel = FALSE
# excel = TRUE, overwrite = TRUE
)## [1] "TADA_DefineCriteriaMethodology: USEPA was included in your 'org_id': Including EPA304a recommended criteria by each unique TADA.CharacteristicName if one is found."
TADA_TableExport(MT.CriteriaMethods.user.supplied2)Review, Save and Re-Use Criteria and Methodology Table
Once the criteria and methodology table has been reviewed and modified, it can be saved and reused for analysis of future TADA data frames. This process is useful to help identify if any new WQP characteristics (or new fraction/speciations) are being queried in your most up to date WQP query and to allow you to determine if an assessment magnitude value should be developed.
For our example, we will use MT.CriteriaMethods_User_Supplied2 and fill in the remaining blank PH magnitude values with a range between 6.5 and 8.5 as MTDEQ criteria of interest.
# We will fill in PH magnitude values for this example
MT.CriteriaMethods.Final <- MT.CriteriaMethods.user.supplied2 |>
dplyr::mutate(MagnitudeValueLower = dplyr::case_when(
grepl("PH_NONE_NONE_NONE", TADA.ComparableDataIdentifier) & ATTAINS.OrganizationIdentifier == "MTDEQ" ~ 6.5,
TRUE ~ MagnitudeValueLower
)) |>
dplyr::mutate(MagnitudeValueUpper = dplyr::case_when(
grepl("PH_NONE_NONE_NONE", TADA.ComparableDataIdentifier) & ATTAINS.OrganizationIdentifier == "MTDEQ" ~ 8.5,
TRUE ~ MagnitudeValueUpper
))
TADA_TableExport(MT.CriteriaMethods.Final)We will supply this final crosswalk table to be reused and validated.
MT.CriteriaMethods.Final2 <- TADA_DefineCriteriaMethodology(
.data = tada.MT.clean,
org_id = "MTDEQ",
MLSummaryRef = NULL,
criteriaMethods = MT.CriteriaMethods.Final,
displayUniqueId = TRUE,
excel = FALSE
# excel = TRUE, overwrite = TRUE
)
TADA_TableExport(MT.CriteriaMethods.Final2)